5FOG
 
 | | Crystal structure of hte Cryptosporidium muris cytosolic leucyl-tRNA synthetase editing domain complex with a post-transfer editing analogue of norvaline (Nv2AA) | | Descriptor: | 1,2-ETHANEDIOL, 2'-(L-NORVALYL)AMINO-2'-DEOXYADENOSINE, LEUCYL-TRNA SYNTHETASE, ... | | Authors: | Palencia, A, Liu, R.J, Lukarska, M, Gut, J, Bougdour, A, Touquet, B, Wang, E.D, Alley, M.R.K, Rosenthal, P.J, Hakimi, M.A, Cusack, S. | | Deposit date: | 2015-11-20 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cryptosporidium and Toxoplasma Parasites are Inhibited by a Benzoxaborole Targeting Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
7QGL
 
 | |
7QGO
 
 | | Human CD73 (ecto 5'-nucleotidase) in complex with MRS4602 (a 3-methyl-CMPCP derivative, compound 21 in paper) in the closed state (crystal form III) | | Descriptor: | 5'-nucleotidase, ZINC ION, [[(2~{R},3~{S},4~{R},5~{R})-5-[(4~{E})-4-[(4-methoxycarbonylphenyl)methoxyimino]-3-methyl-2-oxidanylidene-pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]methylphosphonic acid | | Authors: | Strater, N. | | Deposit date: | 2021-12-09 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structure-Activity Relationship of 3-Methylcytidine-5'-alpha , beta-methylenediphosphates as CD73 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7QGA
 
 | |
5D0G
 
 | |
5D15
 
 | | Crystal structure of an adenylyl cyclase Ma1120 from Mycobacterium avium in complex with ATP and calcium ion | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Bharambe, N.G, Barathy, D.V, Suguna, K. | | Deposit date: | 2015-08-03 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate specificity determinants of class III nucleotidyl cyclases
Febs J., 283, 2016
|
|
5B25
 
 | | Crystal structure of human PDE1B with inhibitor 3 | | Descriptor: | (11R,15S)-4-{[4-(6-fluoropyridin-2-yl)phenyl]methyl}-8-methyl-5-(phenylamino)-1,3,4,8,10-pentaazatetracyclo[7.6.0.02,6.011,15]pentadeca-2,5,9-trien-7-one, Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B, GLYCEROL, ... | | Authors: | Ida, K, Lane, W, Snell, G, Sogabe, S. | | Deposit date: | 2016-01-07 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Potent and Selective Inhibitors of Phosphodiesterase 1 for the Treatment of Cognitive Impairment Associated with Neurodegenerative and Neuropsychiatric Diseases
J.Med.Chem., 59, 2016
|
|
5DBO
 
 | | Crystal structure of the tetrameric eIF2B-beta2-delta2 complex from C. thermophilum | | Descriptor: | Translation initiation factor eIF2b-like protein,Translation initiation factor eIF2b-like protein, Translation initiation factor eif-2b-like protein | | Authors: | Kuhle, B, Ficner, R. | | Deposit date: | 2015-08-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Architecture of the eIF2B regulatory subcomplex and its implications for the regulation of guanine nucleotide exchange on eIF2.
Nucleic Acids Res., 43, 2015
|
|
5D0E
 
 | | Crystal Structure of an adenylyl cyclase Ma1120-Cat in complex with GTP and calcium from Mycobacterium avium | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cyclase, ... | | Authors: | Bharambe, N.G, Barathy, D.V, Suguna, K. | | Deposit date: | 2015-08-03 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Substrate specificity determinants of class III nucleotidyl cyclases
Febs J., 283, 2016
|
|
5D0H
 
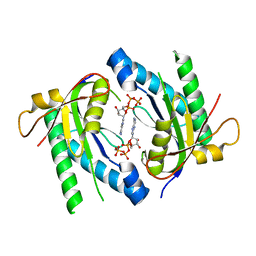 | |
3H86
 
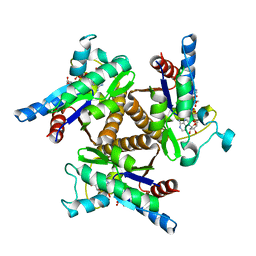 | | Crystal structure of adenylate kinase from Methanococcus maripaludis | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE | | Authors: | Milya, D.G, Yousif, S. | | Deposit date: | 2009-04-28 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a trimeric archaeal adenylate kinase from the mesophile Methanococcus maripaludis with an unusually broad functional range and thermal stability.
Proteins, 78, 2009
|
|
3HID
 
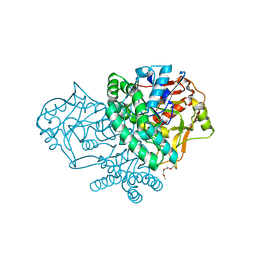 | | Crystal structure of adenylosuccinate synthetase from Yersinia pestis CO92 | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Adenylosuccinate synthetase | | Authors: | Zhang, R, Zhou, M, Peterson, S, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-19 | | Release date: | 2009-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the adenylosuccinate synthetase from Yersinia pestis CO92
To be Published
|
|
3IRW
 
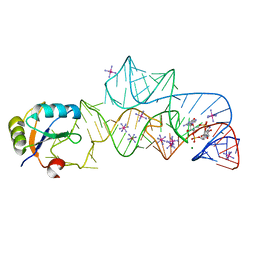 | | Structure of a c-di-GMP riboswitch from V. cholerae | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), IRIDIUM HEXAMMINE ION, MAGNESIUM ION, ... | | Authors: | Smith, K.D. | | Deposit date: | 2009-08-24 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand binding by a c-di-GMP riboswitch.
Nat.Struct.Mol.Biol., 16, 2009
|
|
8FKW
 
 | |
7F0I
 
 | | phosphodiesterase-9A in complex with inhibitor 4b | | Descriptor: | 1-cyclopentyl-6-[[(2R)-1-(6-fluoranyl-2-azaspiro[3.3]heptan-2-yl)-1-oxidanylidene-propan-2-yl]amino]-5H-pyrazolo[3,4-d]pyrimidin-4-one, Isoform PDE9A2 of High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Wu, Y, Huang, Y.Y, Luo, H.B. | | Deposit date: | 2021-06-04 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.70000887 Å) | | Cite: | Discovery of Potent Phosphodiesterase-9 Inhibitors for the Treatment of Hepatic Fibrosis
J.Med.Chem., 64, 2021
|
|
3JVU
 
 | |
3JSL
 
 | |
3JZA
 
 | | Crystal structure of human Rab1b in complex with the GEF domain of DrrA/SidM from Legionella pneumophila | | Descriptor: | PHOSPHATE ION, Ras-related protein Rab-1B, Uncharacterized protein DrrA | | Authors: | Schoebel, S, Oesterlin, L.K, Blankenfeldt, W, Goody, R.S, Itzen, A. | | Deposit date: | 2009-09-23 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RabGDI displacement by DrrA from Legionella is a consequence of its guanine nucleotide exchange activity.
Mol.Cell, 36, 2009
|
|
3JSN
 
 | |
3KHV
 
 | | Crystal Structures of Urokinase-type Plasminogen Activator in Complex with 4-(Aminomethyl) Benzoic Acid and 4-(Aminomethyl-phenyl)-methanol | | Descriptor: | SULFATE ION, TRIETHYLENE GLYCOL, Urokinase-type plasminogen activator, ... | | Authors: | Jiang, L.-G, Zhao, G.-X, Bian, X.-B, Yuan, C, Huang, Z.-X, Huang, M.-D. | | Deposit date: | 2009-10-30 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structures of Urokinase-type Plasminogen Activator in Complex with 4-(Aminomethyl) Benzoic Acid and 4-(Aminomethyl-phenyl)-methanol
CHIN.J.STRUCT.CHEM., 28, 2009
|
|
2J3M
 
 | | PROLYL-TRNA SYNTHETASE FROM ENTEROCOCCUS FAECALIS COMPLEXED WITH ATP, manganese and prolinol | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PROLYL-TRNA SYNTHETASE, ... | | Authors: | Crepin, T, Yaremchuk, A, Tukalo, M, Cusack, S. | | Deposit date: | 2006-08-22 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Two Bacterial Prolyl-tRNA Synthetases with and without a Cis-Editing Domain.
Structure, 14, 2006
|
|
2J3L
 
 | | Prolyl-tRNA synthetase from Enterococcus faecalis complexed with a prolyl-adenylate analogue ('5'-O-(N-(L-PROLYL)-SULFAMOYL)ADENOSINE) | | Descriptor: | '5'-O-(N-(L-PROLYL)-SULFAMOYL)ADENOSINE, PROLYL-TRNA SYNTHETASE, SULFATE ION | | Authors: | Crepin, T, Yaremchuk, A, Tukalo, M, Cusack, S. | | Deposit date: | 2006-08-22 | | Release date: | 2006-10-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Two Bacterial Prolyl-tRNA Synthetases with and without a Cis-Editing Domain.
Structure, 14, 2006
|
|
6EJT
 
 | |
1J70
 
 | | CRYSTAL STRUCTURE OF YEAST ATP SULFURYLASE | | Descriptor: | ATP SULPHURYLASE, PHOSPHATE ION, SODIUM ION | | Authors: | Lalor, D.J, Schnyder, T, Saridakis, V, Pilloff, D.E, Dong, A, Tang, H, Leyh, T.S, Pai, E.F. | | Deposit date: | 2001-05-15 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of a truncated form of Saccharomyces cerevisiae ATP sulfurylase: C-terminal domain essential for oligomer formation but not for activity.
Protein Eng., 16, 2003
|
|
2IEI
 
 | | Crystal structure of rabbit muscle glycogen phosphorylase in complex with 3,4-dihydro-2-quinolone | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, (S)-2-CHLORO-N-(1-(2-(2-HYDROXYETHYLAMINO)-2-OXOETHYL)-2-OXO-1,2,3,4-TETRAHYDROQUINOLIN-3-YL)-6H-THIENO[2,3-B]PYRROLE-5-CARBOXAMIDE, Glycogen phosphorylase, ... | | Authors: | Birch, A.M, Kenny, P.W, Oikonomakos, N.G, Otterbein, L, Schofield, P, Whittamore, P.R.O, Whalley, D.P, Rowsell, S, Pauptit, R, Pannifer, A, Breed, J, Minshull, C. | | Deposit date: | 2006-09-19 | | Release date: | 2006-12-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Development of potent, orally active 1-substituted-3,4-dihydro-2-quinolone glycogen phosphorylase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
