1NBE
 
 | |
3A7Y
 
 | |
3A7Z
 
 | |
3A8B
 
 | |
3A84
 
 | |
2DWK
 
 | | Crystal structure of the RUN domain of mouse Rap2 interacting protein x | | Descriptor: | Protein RUFY3 | | Authors: | Kukimoto-Niino, M, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-15 | | Release date: | 2006-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the RUN Domain of the RAP2-interacting Protein x
J.Biol.Chem., 281, 2006
|
|
3UG3
 
 | | Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase, ... | | Authors: | Im, D.-H, Miyazaki, K, Wakagi, T, Fushinobu, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Glycoside Hydrolase Family 51 alpha-L-Arabinofuranosidase from Thermotoga maritima
Biosci.Biotechnol.Biochem., 76, 2012
|
|
2H3M
 
 | | Crystal Structure of ZO-1 PDZ1 | | Descriptor: | SULFATE ION, Tight junction protein ZO-1 | | Authors: | Appleton, B.A, Zhang, Y, Wu, P, Yin, J.P, Hunziker, W, Skelton, N.J, Sidhu, S.S, Wiesmann, C. | | Deposit date: | 2006-05-22 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Comparative structural analysis of the Erbin PDZ domain and the first PDZ domain of ZO-1. Insights into determinants of PDZ domain specificity.
J.Biol.Chem., 281, 2006
|
|
2KXQ
 
 | | Solution Structure of Smurf2 WW2 and WW3 bound to Smad7 PY motif containing peptide | | Descriptor: | E3 ubiquitin-protein ligase SMURF2, Smad7 PY motif containing peptide | | Authors: | Chong, A, Lin, H, Wrana, J, Forman-Kay, J.D. | | Deposit date: | 2010-05-11 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Coupling of tandem Smad ubiquitination regulatory factor (Smurf) WW domains modulates target specificity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2M9A
 
 | | Solution NMR Structure of E3 ubiquitin-protein ligase ZFP91 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR7784A | | Descriptor: | E3 ubiquitin-protein ligase ZFP91, ZINC ION | | Authors: | Pederson, K, Shastry, R, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-05 | | Release date: | 2013-07-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Hr7784A
To be Published
|
|
6X2U
 
 | | Crystal Structure of PKINES peptide bound to CRM1 | | Descriptor: | Exportin-1, GLYCEROL, GTP-binding nuclear protein Ran, ... | | Authors: | Baumhardt, J.M. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of nuclear export signals by CRM1 carrying the oncogenic E571K mutation.
Mol.Biol.Cell, 31, 2020
|
|
6X4D
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-(cyclopropylmethyl)-7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-8-methyl-2-naphthonitrile (JLJ678), a Non-nucleoside Inhibitor | | Descriptor: | 5-(cyclopropylmethyl)-7-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-8-methylnaphthalene-2-carbonitrile, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
5OL5
 
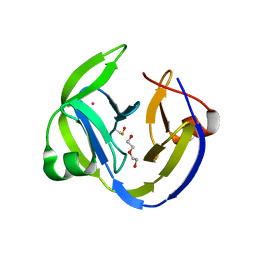 | | Crystal structure of an inactivated Ssp SICLOPPS intein with CFAHPQ extein | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, DNA polymerase III subunit alpha,DNA polymerase III subunit alpha, ... | | Authors: | Kick, L.M, Schneider, S. | | Deposit date: | 2017-07-26 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.329 Å) | | Cite: | Mechanistic Insights into Cyclic Peptide Generation by DnaE Split-Inteins through Quantitative and Structural Investigation.
Chembiochem, 18, 2017
|
|
2M3O
 
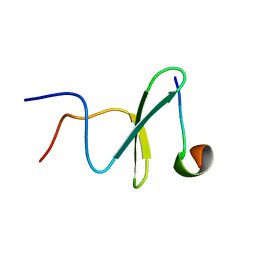 | | Structure and dynamics of a human Nedd4 WW domain-ENaC complex | | Descriptor: | Amiloride-sensitive sodium channel subunit alpha, E3 ubiquitin-protein ligase NEDD4 | | Authors: | Bobby, R, Medini, K, Neudecker, P, Lee, V, MacDonald, F.J, Brimble, M.A, Lott, J, Dingley, A.J. | | Deposit date: | 2013-01-23 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of human Nedd4-1 WW3 in complex with the alpha ENaC PY motif.
Biochim.Biophys.Acta, 1834, 2013
|
|
6X2S
 
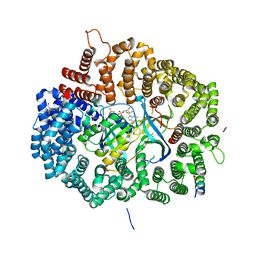 | | Crystal Structure of Mek1(NQ)NES peptide bound to CRM | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, Exportin-1, GLYCEROL, ... | | Authors: | Baumhardt, J.M. | | Deposit date: | 2020-05-20 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | Recognition of nuclear export signals by CRM1 carrying the oncogenic E571K mutation.
Mol.Biol.Cell, 31, 2020
|
|
5OL7
 
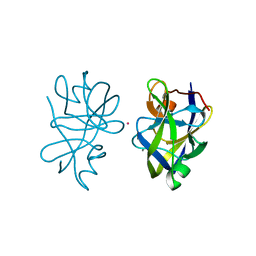 | |
6GFG
 
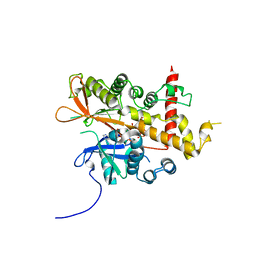 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with D-chiro-IP6 and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-chiro inositol hexakisphosphate, Inositol-pentakisphosphate 2-kinase, ... | | Authors: | Whitfield, H.L, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-04-30 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Fluorescent Probe Identifies Active Site Ligands of Inositol Pentakisphosphate 2-Kinase.
J. Med. Chem., 61, 2018
|
|
6X2P
 
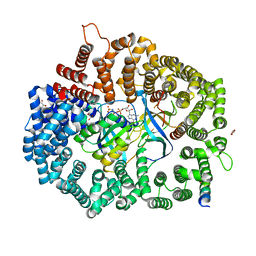 | | Crystal Structure of the Mek1NES peptide bound to CRM1 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, Exportin-1, GLYCEROL, ... | | Authors: | Baumhardt, J.M. | | Deposit date: | 2020-05-20 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Recognition of nuclear export signals by CRM1 carrying the oncogenic E571K mutation.
Mol.Biol.Cell, 31, 2020
|
|
6WJ6
 
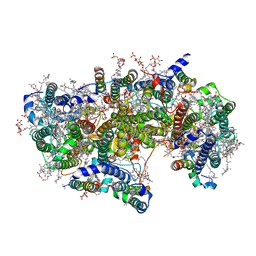 | | Cryo-EM structure of apo-Photosystem II from Synechocystis sp. PCC 6803 | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Gisriel, C.J. | | Deposit date: | 2020-04-12 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Cryo-EM Structure of Monomeric Photosystem II from Synechocystis sp. PCC 6803 Lacking the Water-Oxidation Complex
Joule, 2020
|
|
6X2V
 
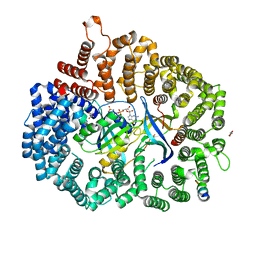 | | Crystal Structure of PKI(DE)NES peptide bound to CRM1 | | Descriptor: | Exportin-1, GLYCEROL, GTP-binding nuclear protein Ran, ... | | Authors: | Baumhardt, J.M. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.822 Å) | | Cite: | Recognition of nuclear export signals by CRM1 carrying the oncogenic E571K mutation.
Mol.Biol.Cell, 31, 2020
|
|
6X49
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with 7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-2-naphthonitrile (JLJ649), a Non-nucleoside Inhibitor | | Descriptor: | 7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-2-naphthonitrile, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.745 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
6X4F
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with methyl 2-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)acetate (JLJ681), a Non-nucleoside Inhibitor | | Descriptor: | Reverse transcriptase/ribonuclease H, SULFATE ION, methyl (6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)acetate, ... | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
6X47
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-2-naphthonitrile (JLJ649), a Non-nucleoside Inhibitor | | Descriptor: | 7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-2-naphthonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.767 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
6X4E
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with methyl 2-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)acetate (JLJ681), a Non-nucleoside Inhibitor | | Descriptor: | Reverse transcriptase/ribonuclease H, SULFATE ION, methyl (6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)acetate, ... | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
6X2Y
 
 | |
