4AEA
 
 | | Dimeric alpha-cobratoxin X-ray structure: Localization of intermolecular disulfides and possible mode of binding to nicotinic acetylcholine receptors | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCINE, LONG NEUROTOXIN 1 | | Authors: | Rucktooa, P, Osipov, A.V, Kasheverov, I.E, Filkin, S.Y, Starkov, V.G, Andreeva, T.V, Bertrand, D, Utkin, Y.N, Tsetlin, V.I, Sixma, T.K. | | Deposit date: | 2012-01-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Dimeric Alpha-Cobratoxin X-Ray Structure: Localization of Intermolecular Disulfides and Possible Mode of Binding to Nicotinic Acetylcholine Receptors.
J.Biol.Chem., 287, 2012
|
|
6T1K
 
 | | Streptavidin variants harbouring an artificial organocatalyst based cofactor | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-(1-pyridin-4-ylpiperidin-4-yl)pentanamide, Streptavidin | | Authors: | Lechner, H, Hocker, B. | | Deposit date: | 2019-10-04 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An Artificial Cofactor Catalyzing the Baylis-Hillman Reaction with Designed Streptavidin as Protein Host*.
Chembiochem, 22, 2021
|
|
3CWU
 
 | | Crystal Structure of an AlkA Host/Guest Complex 2'-fluoro-2'-deoxy-1,N6-ethenoadenine:Thymine Base Pair | | Descriptor: | DNA (5'-D(*DGP*DAP*DCP*DAP*DTP*DGP*DAP*(2FE)P*DTP*DGP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DAP*DTP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3'), DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the E. coli DNA Glycosylase AlkA Bound to the Ends of Duplex DNA: A System for the Structure Determination of Lesion-Containing DNA.
Structure, 16, 2008
|
|
3CXY
 
 | |
3D3Z
 
 | | Crystal structure of Actibind a T2 RNase | | Descriptor: | 1,2-ETHANEDIOL, 1-(5-deoxy-beta-L-xylofuranosyl)pyrimidine-2,4(1H,3H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gonzalez, A, Almog, O. | | Deposit date: | 2008-05-13 | | Release date: | 2009-06-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structue of Actibind a T2 RNase
To be Published
|
|
2N9T
 
 | | NMR solution structure of ProTx-II | | Descriptor: | Beta/omega-theraphotoxin-Tp2a | | Authors: | Schroeder, C.I. | | Deposit date: | 2015-12-08 | | Release date: | 2016-07-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Interaction of Tarantula Venom Peptide ProTx-II with Lipid Membranes Is a Prerequisite for Its Inhibition of Human Voltage-gated Sodium Channel NaV1.7.
J.Biol.Chem., 291, 2016
|
|
6T6V
 
 | | Glu-494-Ala inactive monomer of a quinol dependent Nitric Oxide Reductase (qNOR) from Alcaligenes xylosoxidans | | Descriptor: | CALCIUM ION, Nitric oxide reductase subunit B, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gopalasingam, C.C, Johnson, R.M, Antonyuk, S.V, Muench, S.P, Hasnain, S.S. | | Deposit date: | 2019-10-19 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The active form of quinol-dependent nitric oxide reductase fromNeisseria meningitidisis a dimer.
Iucrj, 7, 2020
|
|
3D17
 
 | | A triply ligated crystal structure of relaxed state human hemoglobin | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Safo, M.K, Musayev, F.N, Jenkins, J, Abraham, D.J. | | Deposit date: | 2008-05-05 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A triply ligated crystal structure of relaxed state human hemoglobin
TO BE PUBLISHED
|
|
3D7O
 
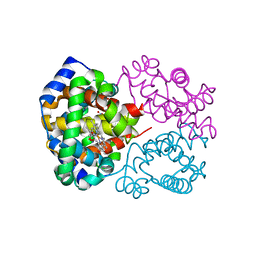 | | Human hemoglobin, nitrogen dioxide anion modified | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, NITRITE ION, ... | | Authors: | Yi, J, Safo, M.K, Richter-Addo, G.B. | | Deposit date: | 2008-05-21 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The nitrite anion binds to human hemoglobin via the uncommon O-nitrito mode.
Biochemistry, 47, 2008
|
|
3D7V
 
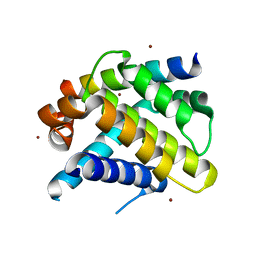 | | Crystal structure of Mcl-1 in complex with an Mcl-1 selective BH3 ligand | | Descriptor: | Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Lee, E.F, Czabotar, P.E, Colman, P.M, Fairlie, W.D. | | Deposit date: | 2008-05-22 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A novel BH3 ligand that selectively targets Mcl-1 reveals that apoptosis can proceed without Mcl-1 degradation.
J.Cell Biol., 180, 2008
|
|
6MX7
 
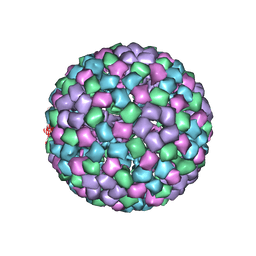 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus: Genome-Binding Capsid N-terminal Domain | | Descriptor: | Capsid | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
3CH9
 
 | |
2N4T
 
 | |
3CHO
 
 | | Crystal structure of leukotriene a4 hydrolase in complex with 2-amino-N-[4-(phenylmethoxy)phenyl]-acetamide | | Descriptor: | ACETATE ION, Leukotriene A-4 hydrolase, N-[4-(benzyloxy)phenyl]glycinamide, ... | | Authors: | Thunnissen, M.M.G.M, Adler, M, Whitlow, M. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis of glutamic acid analogs as potent inhibitors of leukotriene A4 hydrolase.
Bioorg.Med.Chem., 16, 2008
|
|
6MZZ
 
 | | Fluoroacetate dehalogenase, room temperature structure, using first 1 degree of total 3 degree oscillation | | Descriptor: | CALCIUM ION, Fluoroacetate dehalogenase | | Authors: | Finke, A.D, Wierman, J.L, Pare-Labrosse, O, Sarrachini, A, Besaw, J, Mehrabi, P, Gruner, S.M, Miller, R.J.D. | | Deposit date: | 2018-11-06 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fixed-target serial oscillation crystallography at room temperature.
IUCrJ, 6, 2019
|
|
3CKB
 
 | | B. thetaiotaomicron SusD with maltotriose | | Descriptor: | CALCIUM ION, SusD, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Koropatkin, N.M, Martens, E.C, Gordon, J.I, Smith, T.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices.
Structure, 16, 2008
|
|
4A0H
 
 | | Structure of bifunctional DAPA aminotransferase-DTB synthetase from Arabidopsis thaliana bound to 7-keto 8-amino pelargonic acid (KAPA) | | Descriptor: | 7-KETO-8-AMINOPELARGONIC ACID, ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, L(+)-TARTARIC ACID, ... | | Authors: | Cobessi, D, Dumas, R, Pautre, V, Meinguet, C, Ferrer, J.L, Alban, C. | | Deposit date: | 2011-09-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.808 Å) | | Cite: | Biochemical and Structural Characterization of the Arabidopsis Bifunctional Enzyme Dethiobiotin Synthetase-Diaminopelargonic Acid Aminotransferase: Evidence for Substrate Channeling in Biotin Synthesis.
Plant Cell, 24, 2012
|
|
2MYS
 
 | |
6T9S
 
 | | Human Butyrylcholinesterase in complex with 2-(N-hydroxyimino)-N-[(1S)-3-{4-[(2-methyl-1H-imidazol-1-yl)methyl]-1H-1,2,3-triazol-1-yl}-1- phenylpropyl]acetamide | | Descriptor: | (2~{E})-2-hydroxyimino-~{N}-[(1~{S})-3-[4-[(2-methylimidazol-1-yl)methyl]-1,2,3-triazol-1-yl]-1-phenyl-propyl]ethanamid e, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(5-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Sinko, G, Marakovic, N, Knezevic, A. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enantioseparation, in vitro testing, and structural characterization of triple-binding reactivators of organophosphate-inhibited cholinesterases.
Biochem.J., 477, 2020
|
|
2OBI
 
 | |
6N4E
 
 | | hPGDS complexed with a quinoline-3-carboxamide | | Descriptor: | 7-(difluoromethoxy)-N-[trans-4-(2-hydroxypropan-2-yl)cyclohexyl]quinoline-3-carboxamide, GLUTATHIONE, Hematopoietic prostaglandin D synthase | | Authors: | Shewchuk, L.M, Ward, P. | | Deposit date: | 2018-11-19 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The discovery of quinoline-3-carboxamides as hematopoietic prostaglandin D synthase (H-PGDS) inhibitors.
Bioorg. Med. Chem., 27, 2019
|
|
3CY1
 
 | |
6N4Q
 
 | | CryoEM structure of Nav1.7 VSD2 (actived state) in complex with the gating modifier toxin ProTx2 | | Descriptor: | Beta/omega-theraphotoxin-Tp2a, Fab heavy chain, Fab light chain, ... | | Authors: | Xu, H, Rohou, A, Arthur, C.P, Estevez, A, Ciferri, C, Payandeh, J, Koth, C.M. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis of Nav1.7 Inhibition by a Gating-Modifier Spider Toxin.
Cell, 176, 2019
|
|
3CZ7
 
 | |
2NSL
 
 | | E. coli PurE H45N mutant complexed with CAIR | | Descriptor: | 5-AMINO-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, Phosphoribosylaminoimidazole carboxylase catalytic subunit | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-11-04 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N(5)-CAIR Mutase: Role of a CO(2) Binding Site and Substrate Movement in Catalysis.
Biochemistry, 46, 2007
|
|
