4HEU
 
 | |
5I0L
 
 | | Crystal structure of the catalytic domain of MMP-12 in complex with a selective sugar-conjugated arylsulfonamide carboxylate water-soluble inhibitor (DC27). | | Descriptor: | (2R)-2-[{(E)-2-[({(2R,3R,4R,5S,6R)-3-(acetylamino)-4,5-bis(acetyloxy)-6-[(acetyloxy)methyl]tetrahydro-2H-pyran-2-yl}carbamothioyl)amino]ethenyl}(biphenyl-4-ylsulfonyl)amino]-3-methylbutanoic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Stura, E.A, Rosalia, L, Cuffaro, D, Tepshi, L, Ciccone, L, Rossello, A. | | Deposit date: | 2016-02-04 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Sugar-Based Arylsulfonamide Carboxylates as Selective and Water-Soluble Matrix Metalloproteinase-12 Inhibitors.
Chemmedchem, 11, 2016
|
|
4EO8
 
 | | HCV NS5B polymerase inhibitors: Tri-substituted acylhydrazines as tertiary amide bioisosteres | | Descriptor: | 5-(3,3-dimethylbut-1-yn-1-yl)-3-{2,2-dimethyl-1-[(trans-4-methylcyclohexyl)carbonyl]hydrazinyl}thiophene-2-carboxylic acid, RNA-directed RNA polymerase | | Authors: | Appleby, T.C, Canales, E, Watkins, W.J. | | Deposit date: | 2012-04-13 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Tri-substituted acylhydrazines as tertiary amide bioisosteres: HCV NS5B polymerase inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1I8F
 
 | | THE CRYSTAL STRUCTURE OF A HEPTAMERIC ARCHAEAL SM PROTEIN: IMPLICATIONS FOR THE EUKARYOTIC SNRNP CORE | | Descriptor: | GLYCEROL, PUTATIVE SNRNP SM-LIKE PROTEIN | | Authors: | Mura, C, Cascio, D, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2001-03-14 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of a heptameric archaeal Sm protein: Implications for the eukaryotic snRNP core.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1FAZ
 
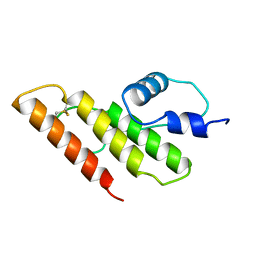 | |
8ZBO
 
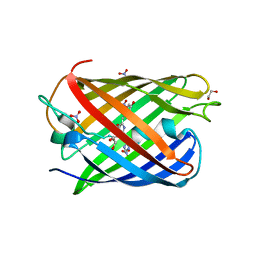 | | Crystal structure of the biphotochromic fluorescent protein moxSAASoti (F97M variant) in its green on-state | | Descriptor: | 1,2-ETHANEDIOL, F97M variant of the biphotochromic fluorescent protein moxSAASoti, NITRATE ION, ... | | Authors: | Boyko, K.M, Matyuta, I.O, Marynich, N.K, Minyaev, M.E, Khadiyatova, A.A, Popov, V.O, Savitsky, A.P. | | Deposit date: | 2024-04-26 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single-point substitution F97M leads to in cellulo crystallization of the biphotochromic protein moxSAASoti.
Biochem.Biophys.Res.Commun., 732, 2024
|
|
4KCB
 
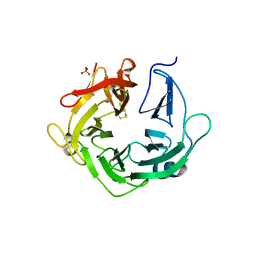 | | Crystal Structure of Exo-1,5-alpha-L-arabinanase from Bovine Ruminal Metagenomic Library | | Descriptor: | Arabinan endo-1,5-alpha-L-arabinosidase, PHOSPHATE ION | | Authors: | Santos, C.R, Polo, C.C, Costa, M.C.M.F, Nascimento, A.F.Z, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-05 | | Last modified: | 2014-04-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases.
J.Biol.Chem., 289, 2014
|
|
4EO6
 
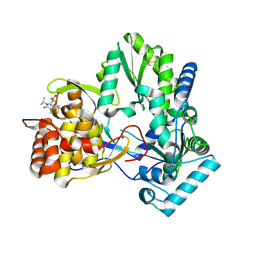 | |
5GMZ
 
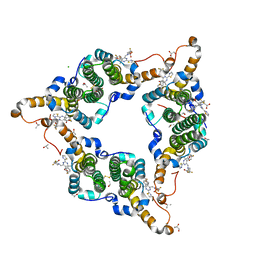 | | Hepatitis B virus core protein Y132A mutant in complex with 4-methyl heteroaryldihydropyrimidine | | Descriptor: | (2S)-4,4-difluoro-1-[[(4S)-4-(4-fluorophenyl)-5-methoxycarbonyl-4-methyl-2-(1,3-thiazol-2-yl)-1H-pyrimidin-6-yl]methyl]pyrrolidine-2-carboxylic acid, CHLORIDE ION, Core protein, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2016-07-18 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Synthesis of Orally Bioavailable 4-Methyl Heteroaryldihydropyrimidine Based Hepatitis B Virus (HBV) Capsid Inhibitors
J.Med.Chem., 59, 2016
|
|
1NGM
 
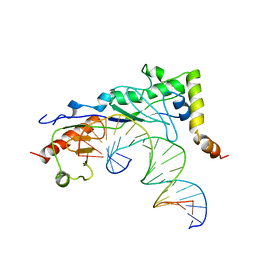 | | Crystal structure of a yeast Brf1-TBP-DNA ternary complex | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*AP*CP*AP*TP*TP*TP*TP*TP*TP*TP*AP*TP*AP*G)-3', 5'-D(*CP*TP*AP*TP*AP*AP*AP*AP*AP*AP*AP*TP*GP*TP*TP*TP*TP*TP*T)-3', Transcription factor IIIB BRF1 subunit, ... | | Authors: | Juo, Z.S, Kassavetis, G.A, Wang, J, Geiduschek, E.P, Sigler, P.B. | | Deposit date: | 2002-12-17 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of a transcription factor IIIB core interface ternary complex
Nature, 422, 2003
|
|
4KCA
 
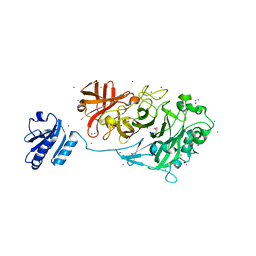 | | Crystal Structure of Endo-1,5-alpha-L-arabinanase from a Bovine Ruminal Metagenomic Library | | Descriptor: | Endo-1,5-alpha-L-arabinanase, GLYCEROL, IODIDE ION, ... | | Authors: | Santos, C.R, Polo, C.C, Costa, M.C.M.F, Nascimento, A.F.Z, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-05 | | Last modified: | 2014-04-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases.
J.Biol.Chem., 289, 2014
|
|
3IYH
 
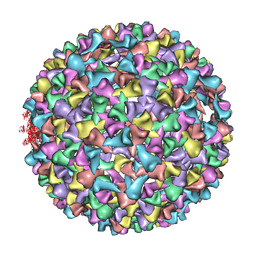 | | P22 procapsid coat protein structures reveal a novel mechanism for capsid maturation: Stability without auxiliary proteins or chemical cross-links | | Descriptor: | Coat protein | | Authors: | Parent, K.N, Khayat, R, Tu, L.H, Suhanovsky, M.M, Cortines, J.R, Teschke, C.M, Johnson, J.E, Baker, T.S. | | Deposit date: | 2009-12-14 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | P22 coat protein structures reveal a novel mechanism for capsid maturation: stability without auxiliary proteins or chemical crosslinks
Structure, 18, 2010
|
|
2LKN
 
 | | Solution structure of the PPIase domain of human aryl-hydrocarbon receptor-interacting protein (AIP) | | Descriptor: | AH receptor-interacting protein | | Authors: | Linnert, M, Lin, Y, Manns, A, Haupt, K, Paschke, A, Fischer, G, Weiwad, M, Luecke, C. | | Deposit date: | 2011-10-17 | | Release date: | 2012-10-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The FKBP-Type Domain of the Human Aryl Hydrocarbon Receptor-Interacting Protein Reveals an Unusual Hsp90 Interaction.
Biochemistry, 52, 2013
|
|
3IYI
 
 | | P22 expanded head coat protein structures reveal a novel mechanism for capsid maturation: Stability without auxiliary proteins or chemical cross-links | | Descriptor: | P22 coat protein in procapsid shells | | Authors: | Parent, K.N, Khayat, R, Tu, L.H, Suhanovsky, M.M, Cortines, J.R, Teschke, C.M, Johnson, J.E, Baker, T.S. | | Deposit date: | 2009-12-14 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | P22 coat protein structures reveal a novel mechanism for capsid maturation: stability without auxiliary proteins or chemical crosslinks
Structure, 18, 2010
|
|
1PXU
 
 | |
1PJZ
 
 | |
4Y30
 
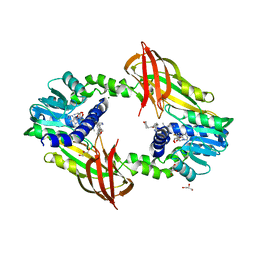 | | Crystal structure of human protein arginine methyltransferase PRMT6 bound to SAH and EPZ020411 | | Descriptor: | GLYCEROL, MAGNESIUM ION, N,N'-dimethyl-N-({3-[4-({trans-3-[2-(tetrahydro-2H-pyran-4-yl)ethoxy]cyclobutyl}oxy)phenyl]-1H-pyrazol-4-yl}methyl)ethane-1,2-diamine, ... | | Authors: | Swinger, K.K, Boriack-Sjodin, P.A. | | Deposit date: | 2015-02-10 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aryl Pyrazoles as Potent Inhibitors of Arginine Methyltransferases: Identification of the First PRMT6 Tool Compound.
Acs Med.Chem.Lett., 6, 2015
|
|
6IHX
 
 | | Crystal Structure Analysis of bovine Hemoglobin modified by SNP | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Kihira, K, Morita, Y, Yamada, T, Kureishi, M, Komatsu, T. | | Deposit date: | 2018-10-03 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Quaternary Structure Analysis of a Hemoglobin Core in Hemoglobin-Albumin Cluster.
J Phys Chem B, 122, 2018
|
|
6JKU
 
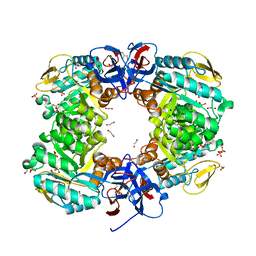 | | Crystal structure of N-acetylglucosamine-6-phosphate deacetylase from Pasteurella Multocida | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-acetylglucosamine-6-phosphate deacetylase, ... | | Authors: | Manjunath, L, Bose, S, Subramanian, R. | | Deposit date: | 2019-03-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Quaternary variations in the structural assembly of N-acetylglucosamine-6-phosphate deacetylase from Pasteurella multocida.
Proteins, 2020
|
|
1T4M
 
 | |
3FW6
 
 | |
4J7A
 
 | | Crystal Structure of Est25 - a Bacterial Homolog of Hormone-Sensitive Lipase from a Metagenomic Library | | Descriptor: | Esterase | | Authors: | Ngo, T.D, Ryu, B.H, Ju, H.S, Jang, E.J, Park, K.S, Joo, S.B, Kim, K.K, Kim, D.H. | | Deposit date: | 2013-02-13 | | Release date: | 2014-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.492 Å) | | Cite: | Structural and functional analyses of a bacterial homologue of hormone-sensitive lipase from a metagenomic library
Acta Crystallogr.,Sect.D, 69, 2014
|
|
3G49
 
 | |
4ZFB
 
 | | Cytochrome P450 pentamutant from BM3 bound to Palmitic Acid | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional P-450/NADPH-P450 reductase, NICKEL (II) ION, ... | | Authors: | Rogers, W.E, Othman, T, Heidary, D.K, Huxford, T. | | Deposit date: | 2015-04-21 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Effect of Mutation and Substrate Binding on the Stability of Cytochrome P450BM3 Variants.
Biochemistry, 55, 2016
|
|
4ZFA
 
 | | Cytochrome P450 wild type from BM3 with bound PEG | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional P-450/NADPH-P450 reductase, NICKEL (II) ION, ... | | Authors: | Rogers, W.E, Othman, T, Heidary, D.K, Huxford, T. | | Deposit date: | 2015-04-21 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.765 Å) | | Cite: | Effect of Mutation and Substrate Binding on the Stability of Cytochrome P450BM3 Variants.
Biochemistry, 55, 2016
|
|
