8A3B
 
 | |
5BK6
 
 | | Structural and biochemical characterization of a non-canonical biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Putative amidase | | Authors: | Peat, T.S, Esquirol, L, Newman, J, Scott, C. | | Deposit date: | 2017-09-12 | | Release date: | 2018-02-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural and biochemical characterization of the biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841.
PLoS ONE, 13, 2018
|
|
6IDS
 
 | | Crystal structure of Vibrio cholerae MATE transporter VcmN D35N mutant | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MATE family efflux transporter | | Authors: | Kusakizako, T, Claxton, D.P, Tanaka, Y, Maturana, A.D, Kuroda, T, Ishitani, R, Mchaourab, H.S, Nureki, O. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Basis of H+-Dependent Conformational Change in a Bacterial MATE Transporter.
Structure, 27, 2019
|
|
6IDR
 
 | | Crystal structure of Vibrio cholerae MATE transporter VcmN in the bent form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MATE family efflux transporter | | Authors: | Kusakizako, T, Claxton, D.P, Tanaka, Y, Maturana, A.D, Kuroda, T, Ishitani, R, Mchaourab, H.S, Nureki, O. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structural Basis of H+-Dependent Conformational Change in a Bacterial MATE Transporter.
Structure, 27, 2019
|
|
4Y2Q
 
 | |
4Y2R
 
 | |
6LLG
 
 | | Crystal Structure of Fagopyrum esculentum M UGT708C1 | | Descriptor: | BENZAMIDINE, SULFATE ION, UDP-glycosyltransferase 708C1 | | Authors: | Wang, X, Liu, M. | | Deposit date: | 2019-12-23 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of theC-Glycosyltransferase UGT708C1 from Buckwheat Provide Insights into the Mechanism ofC-Glycosylation.
Plant Cell, 32, 2020
|
|
6LYY
 
 | | Cryo-EM structure of the human MCT1/Basigin-2 complex in the presence of anti-cancer drug candidate AZD3965 in the outward-open conformation. | | Descriptor: | 3-methyl-5-[[(4~{R})-4-methyl-4-oxidanyl-1,2-oxazolidin-2-yl]carbonyl]-6-[[5-methyl-3-(trifluoromethyl)-1~{H}-pyrazol-4-yl]methyl]-1-propan-2-yl-thieno[2,3-d]pyrimidine-2,4-dione, Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-02-16 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
4Y2X
 
 | | Structure of soluble epoxide hydrolase in complex with 2-({[2-(adamantan-1-yl)ethyl]amino}methyl)phenol | | Descriptor: | 2-[({2-[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]dec-1-yl]ethyl}amino)methyl]phenol, Bifunctional epoxide hydrolase 2, MAGNESIUM ION | | Authors: | Amano, Y, Yamaguchi, T. | | Deposit date: | 2015-02-10 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of N-ethylmethylamine as a novel scaffold for inhibitors of soluble epoxide hydrolase by crystallographic fragment screening
Bioorg.Med.Chem., 23, 2015
|
|
6LRC
 
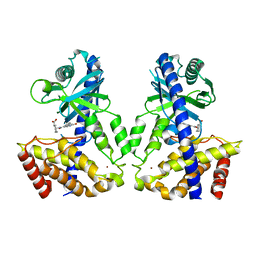 | | Human cGAS catalytic domain bound with the inhibitor PF-06928215 | | Descriptor: | (1R,2S)-2-[(7-hydroxy-5-phenylpyrazolo[1,5-a]pyrimidine-3-carbonyl)amino]cyclohexane-1-carboxylic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Xu, Y.C. | | Deposit date: | 2020-01-15 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.831 Å) | | Cite: | In Silico Screening-Based Discovery of Novel Inhibitors of Human Cyclic GMP-AMP Synthase: A Cross-Validation Study of Molecular Docking and Experimental Testing.
J.Chem.Inf.Model., 60, 2020
|
|
6LRL
 
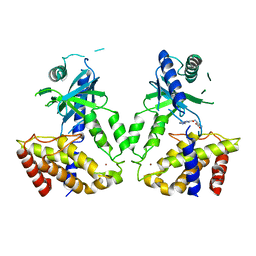 | | Human cGAS catalytic domain bound with compound s2 | | Descriptor: | 3-[[5-(1,2,4-triazol-4-yl)-4H-1,2,4-triazol-3-yl]carbonylamino]benzoic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Xiong, M.Y, Yuan, X.J, Sun, H.B, Xu, Y.C. | | Deposit date: | 2020-01-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.655 Å) | | Cite: | In Silico Screening-Based Discovery of Novel Inhibitors of Human Cyclic GMP-AMP Synthase: A Cross-Validation Study of Molecular Docking and Experimental Testing.
J.Chem.Inf.Model., 60, 2020
|
|
6LLZ
 
 | |
6M67
 
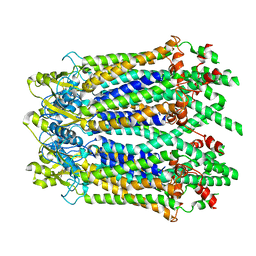 | | The Cryo-EM Structure of Human Pannexin 1 with D376E/D379E Mutation | | Descriptor: | Pannexin-1 | | Authors: | Jin, Q, Bo, Z, Xiang, Z, Xiaokang, Z, Ye, S. | | Deposit date: | 2020-03-13 | | Release date: | 2020-04-15 | | Last modified: | 2020-05-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of human pannexin 1 channel.
Cell Res., 30, 2020
|
|
6LRJ
 
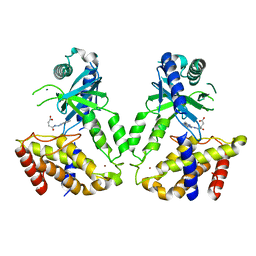 | | Human cGAS catalytic domain bound with compound 23 | | Descriptor: | 4-[2-(2-methyl-[1,2,4]triazolo[1,5-c]quinazolin-5-yl)hydrazinyl]-4-oxidanylidene-butanoic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Xiong, M.Y, Yuan, X.J, Sun, H.B, Xu, Y.C. | | Deposit date: | 2020-01-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | In Silico Screening-Based Discovery of Novel Inhibitors of Human Cyclic GMP-AMP Synthase: A Cross-Validation Study of Molecular Docking and Experimental Testing.
J.Chem.Inf.Model., 60, 2020
|
|
6J79
 
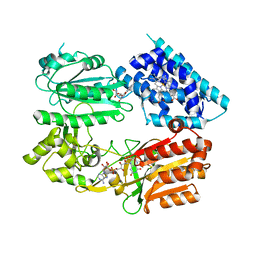 | | Fusion protein of heme oxygenase-1 and NADPH-cytochrome P450 reductase (13aa) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Heme oxygenase 1,NADPH--cytochrome P450 reductase, ... | | Authors: | Sugishima, M, Wada, K. | | Deposit date: | 2019-01-17 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.331 Å) | | Cite: | Crystal structure of a NADPH-cytochrome P450 oxidoreductase (CYPOR) and heme oxygenase 1 fusion protein implies a conformational change in CYPOR upon NADPH/NADP+binding.
Febs Lett., 593, 2019
|
|
4Y2Y
 
 | |
6LLW
 
 | |
4XZE
 
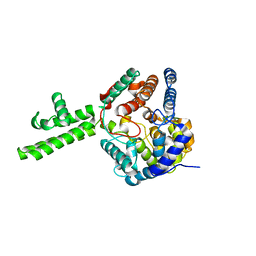 | | The crystal structure of Hazara virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
6LTO
 
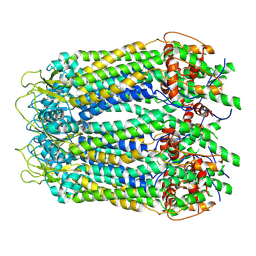 | | cryo-EM structure of full length human Pannexin1 | | Descriptor: | Pannexin-1 | | Authors: | Mou, L.Q, Ke, M, Xiao, Q.J, Wu, J.P, Deng, D. | | Deposit date: | 2020-01-23 | | Release date: | 2020-05-13 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for gating mechanism of Pannexin 1 channel.
Cell Res., 30, 2020
|
|
4Y2U
 
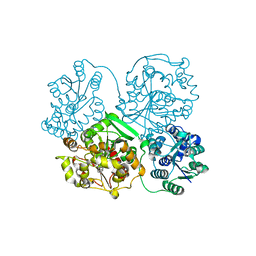 | | Structure of soluble epoxide hydrolase in complex with tert-butyl 1,2,3,4-tetrahydroquinolin-3-ylcarbamate | | Descriptor: | Bifunctional epoxide hydrolase 2, MAGNESIUM ION, tert-butyl (3R)-1,2,3,4-tetrahydroquinolin-3-ylcarbamate | | Authors: | Amano, Y, Yamaguchi, T. | | Deposit date: | 2015-02-10 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification of N-ethylmethylamine as a novel scaffold for inhibitors of soluble epoxide hydrolase by crystallographic fragment screening
Bioorg.Med.Chem., 23, 2015
|
|
3WSU
 
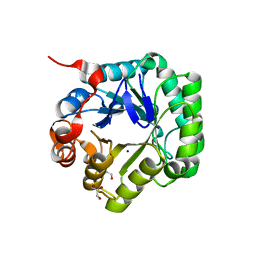 | | Crystal structure of beta-mannanase from Streptomyces thermolilacinus | | Descriptor: | Beta-mannanase, GLYCEROL, SODIUM ION | | Authors: | Kumagai, Y, Yamashita, K, Okuyama, M, Hatanaka, T, Yao, M, Kimura, A. | | Deposit date: | 2014-03-26 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The loop structure of Actinomycete glycoside hydrolase family 5 mannanases governs substrate recognition
Febs J., 282, 2015
|
|
6M68
 
 | | The Cryo-EM Structure of Human Pannexin 1 in the Presence of CBX | | Descriptor: | Pannexin-1 | | Authors: | Jin, Q, Bo, Z, Xiang, Z, Xiaokang, Z, Ye, S. | | Deposit date: | 2020-03-13 | | Release date: | 2020-04-15 | | Last modified: | 2020-05-13 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM structures of human pannexin 1 channel.
Cell Res., 30, 2020
|
|
6M66
 
 | | The Cryo-EM Structure of Human Pannexin 1 | | Descriptor: | Pannexin-1 | | Authors: | Jin, Q, Bo, Z, Xiang, Z, Xiaokang, Z, Ye, S. | | Deposit date: | 2020-03-13 | | Release date: | 2020-04-15 | | Last modified: | 2020-05-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of human pannexin 1 channel.
Cell Res., 30, 2020
|
|
4Y2J
 
 | |
4Y2P
 
 | |
