9C6L
 
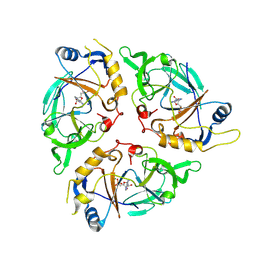 | | Yasminevirus c12orf29, a 5' to 3' RNA ligase | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, RNA ligase1, ... | | Authors: | Hu, Y, Lopez, V.A, Tagliabracci, V.S, Tomchick, D.R. | | Deposit date: | 2024-06-07 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biochemical and structural insights into a 5' to 3' RNA ligase reveal a potential role in tRNA ligation
To Be Published
|
|
4BRE
 
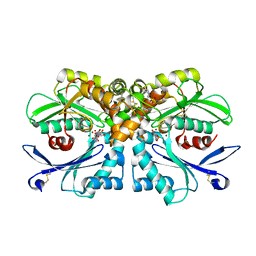 | | Legionella pneumophila NTPDase1 crystal form II (closed) in complex with transition state mimic adenosine 5'phosphovanadate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-PHOSPHOVANADATE, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
5FC5
 
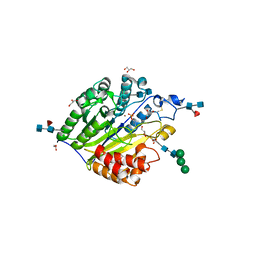 | | Murine SMPDL3A in complex with phosphocholine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3a, ... | | Authors: | Gorelik, A, Illes, K, Superti-Furga, G, Nagar, B. | | Deposit date: | 2015-12-14 | | Release date: | 2016-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.678 Å) | | Cite: | Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A.
J.Biol.Chem., 291, 2016
|
|
5FIT
 
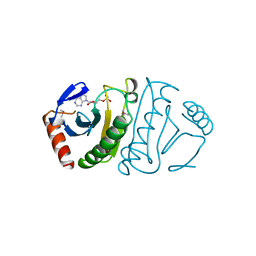 | | FHIT-SUBSTRATE ANALOG | | Descriptor: | FRAGILE HISTIDINE TRIAD PROTEIN, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
5FCA
 
 | | Murine SMPDL3A in presence of excess zinc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3a, ... | | Authors: | Gorelik, A, Illes, K, Superti-Furga, G, Nagar, B. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.924 Å) | | Cite: | Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A.
J.Biol.Chem., 291, 2016
|
|
5FC1
 
 | | Murine SMPDL3A in complex with sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorelik, A, Illes, K, Superti-Furga, G, Nagar, B. | | Deposit date: | 2015-12-14 | | Release date: | 2016-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.389 Å) | | Cite: | Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A.
J.Biol.Chem., 291, 2016
|
|
3IIM
 
 | | The structure of hCINAP-dADP complex at 2.0 angstroms resolution | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Zographos, S.E, Drakou, C.E, Leonidas, D.D. | | Deposit date: | 2009-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | hCINAP is an atypical mammalian nuclear adenylate kinase with an ATPase motif: Structural and functional studies.
Proteins, 80, 2012
|
|
3IIJ
 
 | |
3IIK
 
 | |
5GZ4
 
 | |
3IIL
 
 | | The structure of hCINAP-MgADP-Pi complex at 2.0 angstroms resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Coilin-interacting nuclear ATPase protein, LITHIUM ION, ... | | Authors: | Zographos, S.E, Drakou, C.E, Leonidas, D.D. | | Deposit date: | 2009-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | hCINAP is an atypical mammalian nuclear adenylate kinase with an ATPase motif: Structural and functional studies.
Proteins, 80, 2012
|
|
5FC7
 
 | | Murine SMPDL3A in complex with sulfate (tetragonal) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorelik, A, Illes, K, Superti-Furga, G, Nagar, B. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.456 Å) | | Cite: | Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A.
J.Biol.Chem., 291, 2016
|
|
5HIA
 
 | | Human hypoxanthine-guanine phosphoribosyltransferase in complex with [3R,4R]-4-guanin-9-yl-3-((S)-2-hydroxy-2-phosphonoethyl)oxy-1-N-(phosphonopropionyl)pyrrolidine | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION, [3-[(3~{R},4~{R})-3-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-4-[(2~{S})-2-oxidanyl-2-phosphono-ethoxy]pyrrolidin-1-y l]-3-oxidanylidene-propyl]phosphonic acid | | Authors: | Guddat, L.W, Keough, D.T, Rejman, D. | | Deposit date: | 2016-01-11 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Design of Plasmodium vivax Hypoxanthine-Guanine Phosphoribosyltransferase Inhibitors as Potential Antimalarial Therapeutics.
ACS Chem. Biol., 2017
|
|
5G3Y
 
 | | Crystal structure of adenylate kinase ancestor 1 with Zn and ADP bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENYLATE KINSE, ZINC ION | | Authors: | Nguyen, V, Kutter, S, English, J, Kern, D. | | Deposit date: | 2016-05-03 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Evolutionary drivers of thermoadaptation in enzyme catalysis.
Science, 355, 2017
|
|
5G41
 
 | | Crystal structure of adenylate kinase ancestor 4 with Zn, Mg and Ap5A bound | | Descriptor: | ADENYLATE KINSE, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nguyen, V, Kutter, S, English, J, Kern, D. | | Deposit date: | 2016-05-03 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Evolutionary drivers of thermoadaptation in enzyme catalysis.
Science, 355, 2017
|
|
5HNV
 
 | | Crystal structure of PpkA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Li, P.P, Ran, T.T, Wang, W.W. | | Deposit date: | 2016-01-19 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal structures of the kinase domain of PpkA, a key regulatory component of T6SS, reveal a general inhibitory mechanism.
Biochem.J., 475, 2018
|
|
5GI4
 
 | | DEAD-box RNA helicase | | Descriptor: | ATP-dependent RNA helicase DeaD | | Authors: | Xu, L, Wang, L, Li, F, Wu, L, Shi, Y. | | Deposit date: | 2016-06-22 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Insights into the Structure of Dimeric RNA Helicase CsdA and Indispensable Role of Its C-Terminal Regions.
Structure, 25, 2017
|
|
5G3Z
 
 | | Crystal structure of adenylate kinase ancestor 3 with Zn, Mg and Ap5A bound | | Descriptor: | ADENYLATE KINSE, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nguyen, V, Kutter, S, English, J, Kern, D. | | Deposit date: | 2016-05-03 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Evolutionary drivers of thermoadaptation in enzyme catalysis.
Science, 355, 2017
|
|
1UZU
 
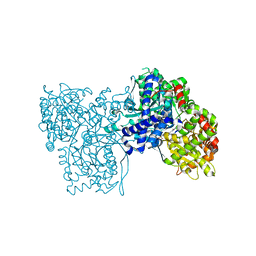 | | Glycogen Phosphorylase b in complex with indirubin-5'-sulphonate | | Descriptor: | 2',3-DIOXO-1,1',2',3-TETRAHYDRO-2,3'-BIINDOLE-5'-SULFONIC ACID, GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, ... | | Authors: | Kosmopoulou, M.N, Leonidas, D.D, Chrysina, E.D, Bischler, N, Eisenbrand, G, Sakarellos, C.E, Pauptit, R, Oikonomakos, N.G. | | Deposit date: | 2004-03-16 | | Release date: | 2004-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding of the potential antitumour agent indirubin-5-sulphonate at the inhibitor site of rabbit muscle glycogen phosphorylase b. Comparison with ligand binding to pCDK2-cyclin A complex.
Eur. J. Biochem., 271, 2004
|
|
2YVN
 
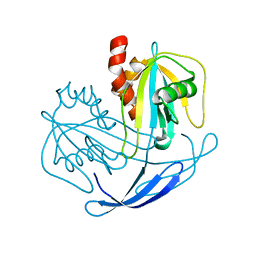 | | Crystal structure of NDX2 from thermus thermophilus HB8 | | Descriptor: | MutT/nudix family protein | | Authors: | Wakamatsu, T, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-13 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for different substrate specificities of two ADP-ribose pyrophosphatases from Thermus thermophilus HB8
J.Bacteriol., 190, 2008
|
|
4BRP
 
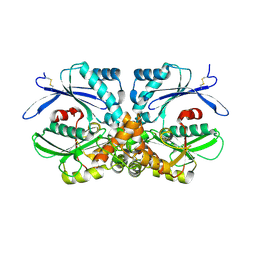 | | Legionella pneumophila NTPDase1 crystal form V (part-open) | | Descriptor: | BROMIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
2YVM
 
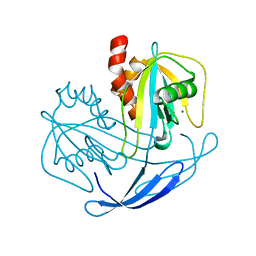 | | Crystal structure of NDX2 in complex with MG2+ from thermus thermophilus HB8 | | Descriptor: | MAGNESIUM ION, MutT/nudix family protein | | Authors: | Wakamatsu, T, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-13 | | Release date: | 2008-02-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for different substrate specificities of two ADP-ribose pyrophosphatases from Thermus thermophilus HB8
J.Bacteriol., 190, 2008
|
|
4I5V
 
 | | Crystal structure of yeast Ap4A phosphorylase Apa2 in complex with Ap4A | | Descriptor: | 5',5'''-P-1,P-4-tetraphosphate phosphorylase 2, BIS(ADENOSINE)-5'-TETRAPHOSPHATE | | Authors: | Jiang, Y.L, Hou, W.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2012-11-29 | | Release date: | 2013-05-08 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | Structures of yeast Apa2 reveal catalytic insights into a canonical AP4A phosphorylase of the histidine triad superfamily
J.Mol.Biol., 425, 2013
|
|
1ADE
 
 | | STRUCTURE OF ADENYLOSUCCINATE SYNTHETASE PH 7 AT 25 DEGREES CELSIUS | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE | | Authors: | Silva, M.M, Poland, B.W, Hoffman, C.M, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1995-09-14 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined crystal structures of unligated adenylosuccinate synthetase from Escherichia coli.
J.Mol.Biol., 254, 1995
|
|
4XYC
 
 | | NANOMOLAR INHIBITORS OF MYCOBACTERIUM TUBERCULOSIS GLUTAMINE SYNTHETASE 1: SYNTHESIS, BIOLOGICAL EVALUATION AND X-RAY CRYSTALLOGRAPHIC STUDIES | | Descriptor: | 9-phenyl-4H-imidazo[1,2-a]indeno[1,2-e]pyrazin-4-one, Glutamine synthetase 1 | | Authors: | Couturier, C, Silve, S, Morales, R, Ppessegue, B, Llopart, S, Nair, A, Bauer, A, Scheiper, B, poeverlein, c, Ganzhorn, A, Lagrange, S, Bacque, E. | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Nanomolar inhibitors of Mycobacterium tuberculosis glutamine synthetase 1: Synthesis, biological evaluation and X-ray crystallographic studies.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
