3TLV
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-22' oxidized mutant in a locally-closed conformation (LC3 subtype) | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-08-30 | | Release date: | 2012-05-16 | | Last modified: | 2012-06-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3TNI
 
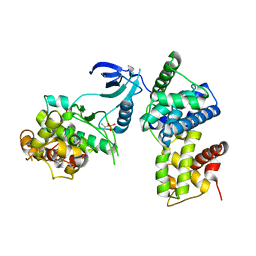 | | structure of CDK9/cyclin T F241L | | Descriptor: | Cyclin-T1, Cyclin-dependent kinase 9 | | Authors: | Baumli, S, Hole, A.J, Endicott, J.A. | | Deposit date: | 2011-09-01 | | Release date: | 2012-02-15 | | Last modified: | 2012-05-30 | | Method: | X-RAY DIFFRACTION (3.234 Å) | | Cite: | The CDK9 C-helix Exhibits Conformational Plasticity That May Explain the Selectivity of CAN508.
Acs Chem.Biol., 7, 2012
|
|
3UAJ
 
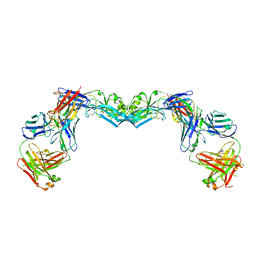 | | Crystal structure of the envelope glycoprotein ectodomain from dengue virus serotype 4 in complex with the fab fragment of the chimpanzee monoclonal antibody 5H2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, monoclonal antibody 5H2, ... | | Authors: | Cockburn, J.J.B, Stura, E.A, Navarro-Sanchez, M.E, Rey, F.A. | | Deposit date: | 2011-10-21 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.232 Å) | | Cite: | Structural insights into the neutralization mechanism of a higher primate antibody against dengue virus.
Embo J., 31, 2012
|
|
3TPW
 
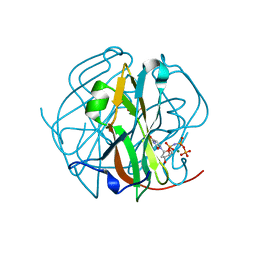 | | CRYSTAL STRUCTURE OF M-PMV DUTPASE - DUPNPP complex revealing distorted ligand geometry (approach intermediate) | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDO HYDROLASE, ... | | Authors: | Barabas, O, Nemeth, V, Vertessy, B.G. | | Deposit date: | 2011-09-08 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Snapshots of Enzyme-Catalysed Phosphate Ester Hydrolysis Directly Visualize In-line Attack and Inversion
to be published
|
|
3TQS
 
 | | Structure of the dimethyladenosine transferase (ksgA) from Coxiella burnetii | | Descriptor: | Ribosomal RNA small subunit methyltransferase A | | Authors: | Rudolph, M, Cheung, J, Franklin, M.C, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2016-02-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TOM
 
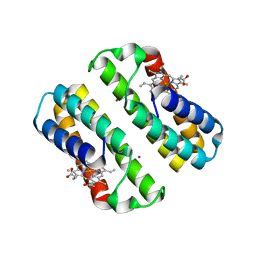 | | Crystal structure of an engineered cytochrome cb562 that forms 2D, Zn-mediated sheets | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Soluble cytochrome b562, ZINC ION | | Authors: | Brodin, J.B, Tezcan, F.A. | | Deposit date: | 2011-09-05 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Metal-directed, chemically tunable assembly of one-, two- and three-dimensional crystalline protein arrays.
Nat Chem, 4, 2012
|
|
3TQY
 
 | | Structure of a single-stranded DNA-binding protein (ssb), from Coxiella burnetii | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.6001 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TR7
 
 | | Structure of a uracil-DNA glycosylase (ung) from Coxiella burnetii | | Descriptor: | Uracil-DNA glycosylase | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1958 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TT8
 
 | |
3TRF
 
 | | Structure of a shikimate kinase (aroK) from Coxiella burnetii | | Descriptor: | SULFATE ION, Shikimate kinase | | Authors: | Cheung, J, Franklin, M, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TRV
 
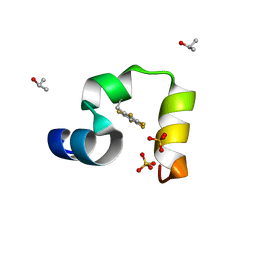 | | Crystal structure of quasiracemic villin headpiece subdomain containing (F5Phe17) substitution | | Descriptor: | D-Villin-1, ISOPROPYL ALCOHOL, L-Villin-1, ... | | Authors: | Mortenson, D.E, Satyshur, K.A, Gellman, S.H, Forest, K.T. | | Deposit date: | 2011-09-11 | | Release date: | 2012-01-25 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Quasiracemic crystallization as a tool to assess the accommodation of noncanonical residues in nativelike protein conformations.
J.Am.Chem.Soc., 134, 2012
|
|
3TRY
 
 | | Crystal structure of racemic villin headpiece subdomain in space group I-4c2 | | Descriptor: | D-Villin-1, SULFATE ION | | Authors: | Mortenson, D.E, Satyshur, K.A, Gellman, S.H, Forest, K.T. | | Deposit date: | 2011-09-11 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Quasiracemic crystallization as a tool to assess the accommodation of noncanonical residues in nativelike protein conformations.
J.Am.Chem.Soc., 134, 2012
|
|
3TVW
 
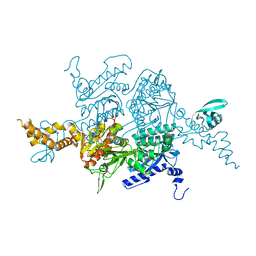 | | Crystal Structure of the humanized carboxyltransferase domain of yeast Acetyl-coA caroxylase in complex with compound 4 | | Descriptor: | Acetyl-CoA carboxylase, [4-(2H-chromen-3-ylmethyl)piperazin-1-yl]-[3-(1H-pyrazol-5-yl)phenyl]methanone | | Authors: | Rajamohan, F, Marr, E, Reyes, A, Landro, J.A, Anderson, M.D, Corbett, J.W, Dirico, K.J, Harwood, J.H, Tu, M, Vajdos, F.F. | | Deposit date: | 2011-09-20 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-guided Inhibitor Design for Human Acetyl-coenzyme A Carboxylase by Interspecies Active Site Conversion.
J.Biol.Chem., 286, 2011
|
|
3TTD
 
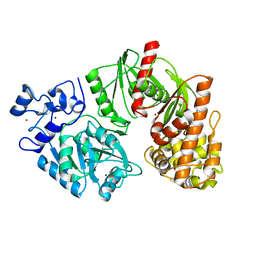 | | Crystal structure of E. coli HypF with AMP-CPP and carbamoyl phosphate | | Descriptor: | MAGNESIUM ION, Transcriptional regulatory protein, ZINC ION | | Authors: | Petkun, S, Shi, R, Li, Y, Cygler, M. | | Deposit date: | 2011-09-14 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Hydrogenase Maturation Protein HypF with Reaction Intermediates Shows Two Active Sites.
Structure, 19, 2011
|
|
3W43
 
 | | Crystal structure of RsbX in complex with manganese in space group P21 | | Descriptor: | MANGANESE (II) ION, Phosphoserine phosphatase RsbX | | Authors: | Teh, A.H, Makino, M, Baba, S, Shimizu, N, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2013-01-04 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structure of the RsbX phosphatase involved in the general stress response of Bacillus subtilis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3W45
 
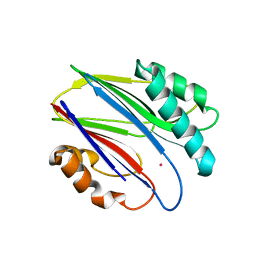 | | Crystal structure of RsbX in complex with cobalt in space group P1 | | Descriptor: | COBALT (II) ION, Phosphoserine phosphatase RsbX | | Authors: | Makino, M, Teh, A.H, Baba, S, Shimizu, N, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2013-01-04 | | Release date: | 2014-01-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the RsbX phosphatase involved in the general stress response of Bacillus subtilis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3W57
 
 | | Structure of a C2 domain | | Descriptor: | C2 domain protein, CALCIUM ION | | Authors: | Traore, D.A.K, Whisstock, J.C. | | Deposit date: | 2013-01-24 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | Defining the interaction of perforin with calcium and the phospholipid membrane.
Biochem.J., 456, 2013
|
|
3V9O
 
 | |
3VCE
 
 | | Thaumatin by LB based Hanging Drop Vapour Diffusion after 18.1 MGy X-Ray dose at ESRF ID29 beamline (Best Case) | | Descriptor: | GLYCEROL, Thaumatin I | | Authors: | Belmonte, L, Pechkova, E, Scudieri, D, Nicolini, C. | | Deposit date: | 2012-01-04 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Langmuir-blodgett nanotemplate and radiation resistance in protein crystals: state of the art.
Crit Rev Eukaryot Gene Expr, 22, 2012
|
|
3VCJ
 
 | | Thaumatin by LB Hanging Drop Vapour Diffusion after 9.05 MGy X-Ray dose at ESRF ID29 beamline (Best Case) | | Descriptor: | GLYCEROL, Thaumatin I | | Authors: | Belmonte, L, Pechkova, E, Scudieri, D, Nicolini, C. | | Deposit date: | 2012-01-04 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Langmuir-blodgett nanotemplate and radiation resistance in protein crystals: state of the art.
Crit Rev Eukaryot Gene Expr, 22, 2012
|
|
3V9K
 
 | |
3VF1
 
 | | Structure of a calcium-dependent 11R-lipoxygenase suggests a mechanism for Ca-regulation | | Descriptor: | 11R-lipoxygenase, FE (II) ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Eek, P, Jarving, R, Jarving, I, Gilbert, N.C, Newcomer, M.E, Samel, N. | | Deposit date: | 2012-01-09 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | Structure of a Calcium-dependent 11R-Lipoxygenase Suggests a Mechanism for Ca2+ Regulation.
J.Biol.Chem., 287, 2012
|
|
3VGW
 
 | | Crystal structure of monoAc-biotin-avidin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(3aS,4S,6aR)-1-acetyl-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoic acid, Avidin, ... | | Authors: | Terai, T, Maki, E, Sugiyama, S, Takahashi, Y, Matsumura, H, Mori, Y, Nagano, T. | | Deposit date: | 2011-08-21 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rational development of caged-biotin protein-labeling agents and some applications in live cells
Chem.Biol., 18, 2011
|
|
3VBE
 
 | |
3VF2
 
 | | Crystal structure of the O-carbamoyltransferase TobZ M473I variant in complex with carbamoyl phosphate and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FE (II) ION, O-carbamoyltransferase TobZ, ... | | Authors: | Parthier, C, Stubbs, M.T, Goerlich, S, Jaenecke, F. | | Deposit date: | 2012-01-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The O-Carbamoyltransferase TobZ Catalyzes an Ancient Enzymatic Reaction.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
