5A13
 
 | |
6NEN
 
 | | Catalytic domain of Proteus mirabilis ScsC | | Descriptor: | Copper resistance protein | | Authors: | Kurth, F, Furlong, E.J, Premkumar, L, Martin, J.L. | | Deposit date: | 2018-12-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Engineered variants provide new insight into the structural properties important for activity of the highly dynamic, trimeric protein disulfide isomerase ScsC from Proteus mirabilis.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7U1C
 
 | | Structure of EstG crystalized with SO4 and Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase domain-containing protein, SODIUM ION, ... | | Authors: | Gabelli, S.B, Chen, Z. | | Deposit date: | 2022-02-20 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | EstG is a novel esterase required for cell envelope integrity in Caulobacter.
Curr.Biol., 33, 2023
|
|
7U59
 
 | |
7U61
 
 | | Crystal Structure of Anti-Nicotine Antibody NIC311 Fab Complexed with Nicotine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, NIC311 Fab Heavy Chain, NIC311 Fab Light Chain, ... | | Authors: | Rodarte, J.V, Pancera, M.P, Liban, T.L. | | Deposit date: | 2022-03-03 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of drug-specific monoclonal antibodies bound to opioids and nicotine reveal a common mode of binding.
Structure, 31, 2023
|
|
4ZY0
 
 | |
8QXP
 
 | |
6Q0R
 
 | | Structure of DDB1-DDA1-DCAF15 complex bound to E7820 and RBM39 | | Descriptor: | 3-cyano-N-(3-cyano-4-methyl-1H-indol-7-yl)benzene-1-sulfonamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Faust, T, Yoon, H, Nowak, R.P, Donovan, K.A, Li, Z, Cai, Q, Eleuteri, N.A, Zhang, T, Gray, N.S, Fischer, E.S. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15.
Nat.Chem.Biol., 16, 2020
|
|
6VW7
 
 | | Formate Dehydrogenase FdsABG subcomplex FdsBG from C. necator - NADH bound | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Young, T. | | Deposit date: | 2020-02-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and kinetic analyses of the FdsBG subcomplex of the cytosolic formate dehydrogenase FdsABG fromCupriavidus necator.
J.Biol.Chem., 295, 2020
|
|
7NYE
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-134 | | Descriptor: | 14-3-3 protein sigma, 4-(4-propan-2-ylpiperazin-1-yl)sulfonylbenzaldehyde, GLYCEROL, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-03-22 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
6Q3Z
 
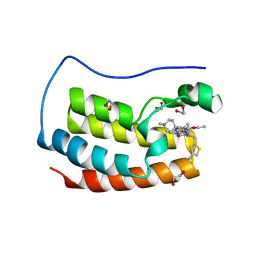 | | Crystal structure of the first bromodomain of human BRD4 in complex with the inhibitor 16k | | Descriptor: | (7~{R})-2-[[2-ethoxy-4-(1-methylpiperidin-4-yl)phenyl]amino]-7-ethyl-5-methyl-8-[(4-methylthiophen-2-yl)methyl]-7~{H}-pteridin-6-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Heidenreich, D, Watts, E, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Knapp, S, Hoelder, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Designing Dual Inhibitors of Anaplastic Lymphoma Kinase (ALK) and Bromodomain-4 (BRD4) by Tuning Kinase Selectivity.
J.Med.Chem., 62, 2019
|
|
7NWS
 
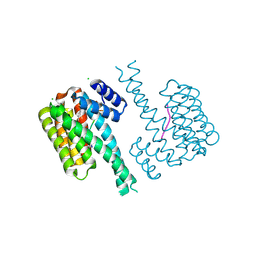 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-185 | | Descriptor: | 14-3-3 protein sigma, 4-[(7-methoxy-2,3-dihydro-1,4-benzoxazin-4-yl)sulfonyl]benzaldehyde, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-03-17 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7TND
 
 | |
8R5D
 
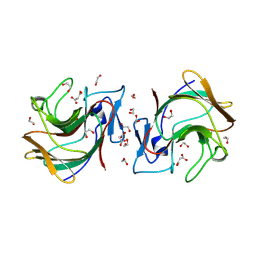 | | Crystal structure of human TRIM7 PRYSPRY domain | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM7, MALONIC ACID | | Authors: | Munoz Sosa, C.J, Kraemer, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-11-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A C-Degron Structure-Based Approach for the Development of Ligands Targeting the E3 Ligase TRIM7.
Acs Chem.Biol., 19, 2024
|
|
6N3K
 
 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 1 | | Descriptor: | Epoxide hydrolase, N-{cis-4-[(2,6-difluorophenyl)methoxy]cyclohexyl}-N'-(3-phenylpropyl)urea | | Authors: | de Oliveira, G.S, Adriani, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors.
Int. J. Biol. Macromol., 129, 2019
|
|
7W43
 
 | |
7NYG
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-127 | | Descriptor: | 14-3-3 protein sigma, 4-[(2~{S})-2-methylpyrrolidin-1-yl]sulfonylbenzaldehyde, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-03-22 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
8RNW
 
 | | Hen Egg White Lysozyme soaked with trans-Ru(DMSO)4Cl2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oszajca, M, Flejszar, M, Szura, A, Drozdz, P, Brindell, M, Kurpiewska, K. | | Deposit date: | 2024-01-11 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Exploring the coordination chemistry of ruthenium complexes with lysozymes: structural and in-solution studies.
Front Chem, 12, 2024
|
|
6BV6
 
 | | Structure of proteinaceous RNase P 1 (PRORP1) from A. thaliana after 3-hour soak with juglone | | Descriptor: | 5-hydroxynaphthalene-1,4-dione, Proteinaceous RNase P 1, chloroplastic/mitochondrial, ... | | Authors: | Karasik, A, Wu, N, Fierke, C.A, Koutmos, M. | | Deposit date: | 2017-12-12 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibition of protein-only RNase P with Gambogic acid and Juglone
To Be Published
|
|
6Q43
 
 | | Atomic resolution crystal structure of an ABA collagen heterotrimer | | Descriptor: | GLYCEROL, Leading Chain of the ABA collagen heterotrimer, Middle Chain of the ABA collagen heterotrimer, ... | | Authors: | Jalan, A.A, Hartgerink, J.D, Brear, P, Leitinger, B, Farndale, R.W. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Atomic resolution crystal structure of an AAB collagen heterotrimer
Nat.Chem.Biol., 2019
|
|
8QLD
 
 | | Bacteriophage T5 dUTPase mutant with loop deletion (30-35 aa) | | Descriptor: | CHLORIDE ION, Deoxyuridine 5'-triphosphate nucleotidohydrolase, GLYCEROL, ... | | Authors: | Gabdulkhakov, A.G, Dzhus, U.F, Selikhanov, G.K, Glukhov, A.S. | | Deposit date: | 2023-09-19 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacteriophage T5 dUTPase: Combination of Common Enzymatic and Novel Functions.
Int J Mol Sci, 25, 2024
|
|
7NXW
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-182 | | Descriptor: | 14-3-3 protein sigma, 4-[(6-chloranyl-3,4-dihydro-2~{H}-quinolin-1-yl)sulfonyl]benzaldehyde, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-03-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
6BVC
 
 | | Crystal structure of AAC(3)-Ia in complex with coenzyme A | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Aminoglycoside-(3)-N-acetyltransferase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Savchenko, A, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-12 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.808 Å) | | Cite: | To be published
To Be Published
|
|
7TNF
 
 | |
7O3A
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-046 | | Descriptor: | 1-[4-(hydroxymethyl)phenyl]sulfonylpiperidin-4-ol, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-04-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
