6KUV
 
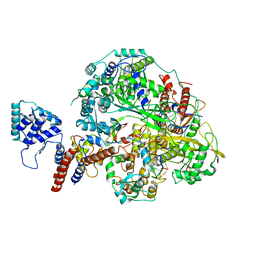 | | Structure of influenza D virus polymerase bound to cRNA promoter in class 2 | | Descriptor: | 3'-cRNA, 5'-cRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUT
 
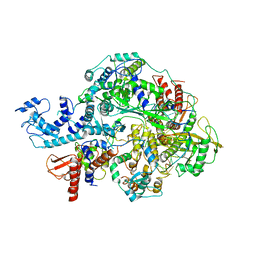 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B2) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6RID
 
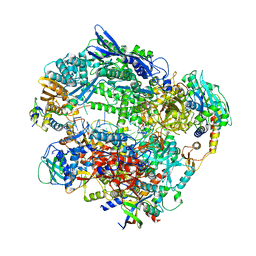 | | Structure of Vaccinia Virus DNA-dependent RNA polymerase elongation complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Hillen, H.S, Bartuli, J, Grimm, C, Dienemann, C, Bedenk, K, Szalar, A, Fischer, U, Cramer, P. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-18 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Transcribing and Capping Vaccinia Complexes.
Cell, 179, 2019
|
|
1MWH
 
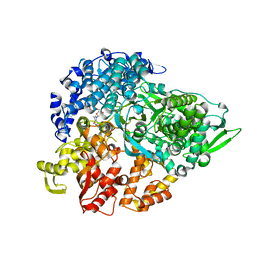 | | REOVIRUS POLYMERASE LAMBDA3 BOUND TO MRNA CAP ANALOG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, MANGANESE (II) ION, MINOR CORE PROTEIN LAMBDA 3 | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-09-29 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA Synthesis in a Cage-Structural Studies of Reovirus Polymerase lambda3
Cell(Cambridge,Mass.), 111, 2002
|
|
6N62
 
 | | Escherichia coli RNA polymerase sigma70-holoenzyme bound to upstream fork promoter DNA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Braffman, N, Hauver, J, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-24 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.803 Å) | | Cite: | Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6N4C
 
 | | EM structure of the DNA wrapping in bacterial open transcription initiation complex | | Descriptor: | DNA (94-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Florez-Ariza, A, Cassago, A, de Oliveira, P.S.L, Guerra, D.G, Portugal, R.V. | | Deposit date: | 2018-11-19 | | Release date: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Interactions of Upstream and Downstream Promoter Regions with RNA Polymerase are Energetically Coupled and a Target of Regulation in Transcription Initiation
Biorxiv, 2020
|
|
6N61
 
 | | Escherichia coli RNA polymerase sigma70-holoenzyme bound to upstream fork promoter DNA and Capistruin | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Capistruin, ... | | Authors: | Braffman, N, Hauver, J, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-24 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.253 Å) | | Cite: | Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6N60
 
 | | Escherichia coli RNA polymerase sigma70-holoenzyme bound to upstream fork promoter DNA and Microcin J25 (MccJ25) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Braffman, N, Hauver, J, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-23 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.68 Å) | | Cite: | Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6M6A
 
 | | Cryo-EM structure of Thermus thermophilus Mfd in complex with RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
6M6C
 
 | | CryoEM structure of Thermus thermophilus RNA polymerase elongation complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
4ZH3
 
 | |
4ZH4
 
 | | Crystal structure of Escherichia coli RNA polymerase in complex with CBRP18 | | Descriptor: | 5-(4-fluorophenyl)-4-[4-fluoro-3-(trifluoromethyl)phenyl]-1H-pyrazole, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Feng, Y, Ebright, R.H. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.993 Å) | | Cite: | Structural Basis of Transcription Inhibition by CBR Hydroxamidines and CBR Pyrazoles.
Structure, 23, 2015
|
|
8Z97
 
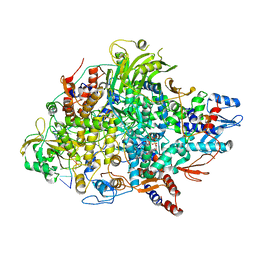 | | Cryo-EM structure of Thogoto virus polymerase in a transcription elongation conformation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z9H
 
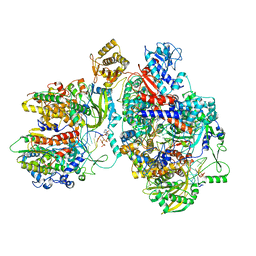 | | Cryo-EM structure of Thogoto virus polymerase in a transcription elongation-reception conformation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
4ZH2
 
 | |
8XMD
 
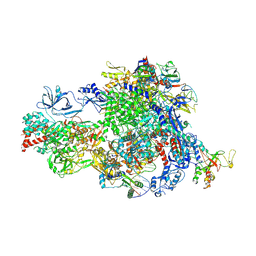 | | Pre-translocated Pol IV transcription elongation complex | | Descriptor: | DNA-directed RNA polymerase IV subunit 1, DNA-directed RNA polymerase IV subunit 7, DNA-directed RNA polymerases II and IV subunit 5A, ... | | Authors: | Huang, K, Fang, C.L, Zhang, Y. | | Deposit date: | 2023-12-27 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Transcription of the Plant RNA polymerase IV is prone to backtracking
To Be Published
|
|
8XME
 
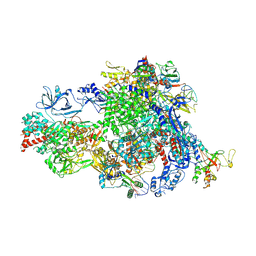 | | Backtracked Pol IV transcription elongation complex | | Descriptor: | DNA-directed RNA polymerase IV subunit 1, DNA-directed RNA polymerase IV subunit 7, DNA-directed RNA polymerases II and IV subunit 5A, ... | | Authors: | Huang, K, Fang, C.L, Zhang, Y. | | Deposit date: | 2023-12-27 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Transcription of the Plant RNA polymerase IV is prone to backtracking
To Be Published
|
|
8XMB
 
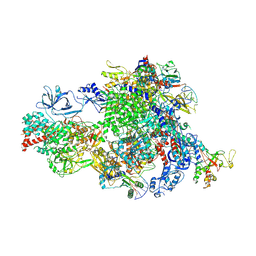 | | NTP-bound Pol IV transcription elongation complex | | Descriptor: | DNA-directed RNA polymerase IV subunit 1, DNA-directed RNA polymerase IV subunit 7, DNA-directed RNA polymerases II and IV subunit 5A, ... | | Authors: | Huang, K, Fang, C.L, Zhang, Y. | | Deposit date: | 2023-12-27 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Transcription of the Plant RNA polymerase IV is prone to backtracking
To Be Published
|
|
8XMC
 
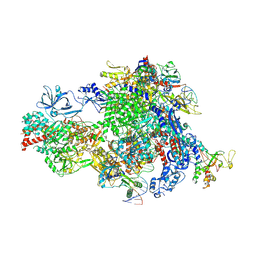 | |
8Z90
 
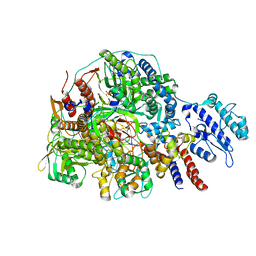 | | Cryo-EM structure of Thogoto virus polymerase in transcription initiation conformation 2 | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z98
 
 | | Cryo-EM structure of Thogoto virus polymerase in a transcription reception conformation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
4ZRL
 
 | | Structure of the non canonical Poly(A) polymerase complex GLD-2 - GLD-3 | | Descriptor: | CHLORIDE ION, Defective in germ line development protein 3, Poly(A) RNA polymerase gld-2 | | Authors: | Nakel, K, Bonneau, F, Eckmann, C.R, Conti, E. | | Deposit date: | 2015-05-12 | | Release date: | 2015-07-08 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis for the activation of the C. elegans noncanonical cytoplasmic poly(A)-polymerase GLD-2 by GLD-3.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8Z8X
 
 | | Cryo-EM structure of Thogoto virus polymerase in a transcription initiation conformation | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
5FMZ
 
 | | Crystal structure of Influenza B polymerase with bound 5' vRNA | | Descriptor: | 5'-R(*AP*GP*UP*AP*GP*UP*AP*AP*CP*AP*AP*GP)-3', POLYMERASE ACIDIC PROTEIN, POLYMERASE BASIC PROTEIN 2, ... | | Authors: | Guilligay, D, Cusack, S. | | Deposit date: | 2015-11-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of Pb2 Domains.
Mol.Cell, 61, 2016
|
|
8PBL
 
 | |
