3U22
 
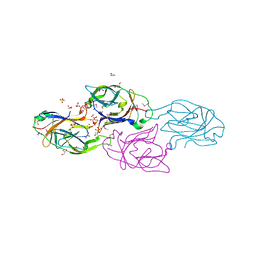 | |
3LGE
 
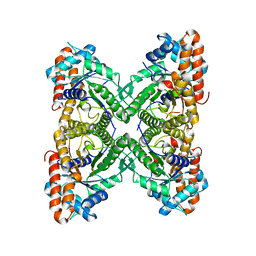 | | Crystal structure of rabbit muscle aldolase-SNX9 LC4 complex | | Descriptor: | Fructose-bisphosphate aldolase A, Sorting nexin-9 | | Authors: | Rangarajan, E.S, Park, H, Fortin, E, Sygusch, J, Izard, T. | | Deposit date: | 2010-01-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of aldolase control of sorting nexin 9 function in endocytosis.
J.Biol.Chem., 285, 2010
|
|
3LP2
 
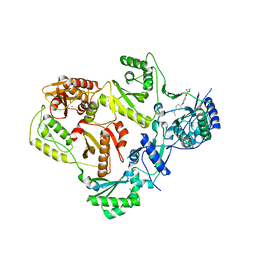 | | HIV-1 reverse transcriptase with inhibitor | | Descriptor: | 3-[4-(diethylamino)phenoxy]-6-(ethoxycarbonyl)-5,8-dihydroxy-7-oxo-7,8-dihydro-1,8-naphthyridin-1-ium, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Yan, Y, Munshi, S.K, Prasad, G.S, Su, H.P. | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors.
J.Virol., 84, 2010
|
|
3LPH
 
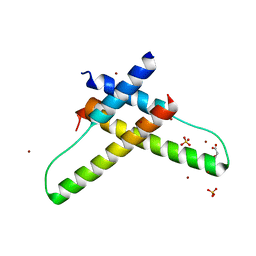 | | Crystal structure of the HIV-1 Rev dimer | | Descriptor: | BROMIDE ION, MALONATE ION, Protein Rev, ... | | Authors: | Daugherty, M.D. | | Deposit date: | 2010-02-05 | | Release date: | 2010-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for cooperative RNA binding and export complex assembly by HIV Rev.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3U3O
 
 | | Crystal structure of Human SULT1A1 bound to PAP and two 3-Cyano-7-hydroxycoumarin | | Descriptor: | 7-hydroxy-2-oxo-2H-chromene-3-carbonitrile, ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase 1A1 | | Authors: | Guttman, C, Berger, I, Aharoni, A, Zarivach, R. | | Deposit date: | 2011-10-06 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The molecular basis for the broad substrate specificity of human sulfotransferase 1A1.
Plos One, 6, 2011
|
|
3LJ2
 
 | | IRE1 complexed with JAK Inhibitor I | | Descriptor: | 2-TERT-BUTYL-9-FLUORO-3,6-DIHYDRO-7H-BENZ[H]-IMIDAZ[4,5-F]ISOQUINOLINE-7-ONE, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Lee, K.P.K, Sicheri, F. | | Deposit date: | 2010-01-25 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1.
Mol.Cell, 38, 2010
|
|
3U4K
 
 | |
3LC1
 
 | | Crystal Structure of H178N mutant of Glyceraldehyde-3-phosphate-dehydrogenase 1 (GAPDH 1) from Staphylococcus aureus MRSA252 complexed with NAD at 2.0 angstrom resolution. | | Descriptor: | GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2010-01-09 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
3U4S
 
 | | Histone Lysine demethylase JMJD2A in complex with T11C peptide substrate crosslinked to N-oxalyl-D-cysteine | | Descriptor: | HISTONE 3 TAIL ANALOG (T11C Peptide), Lysine-specific demethylase 4A, N-(carboxycarbonyl)-D-cysteine, ... | | Authors: | Ma, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2011-10-10 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Linking of 2-Oxoglutarate and Substrate Binding Sites Enables Potent and Highly Selective Inhibition of JmjC Histone Demethylases.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3LCR
 
 | | Thioesterase from Tautomycetin Biosynthhetic Pathway | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, Tautomycetin biosynthetic PKS | | Authors: | Akey, D.L, Scaglione, J.B, Smith, J.L, Sherman, D.H. | | Deposit date: | 2010-01-11 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural characterization of the tautomycetin thioesterase: analysis of a stereoselective polyketide hydrolase.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3LKP
 
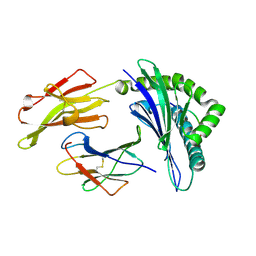 | | Crystal Structure of HLA B*3501 in complex with influenza NP418 epitope from 1972 strain | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Gras, S, Kedzierski, L, Valkenburg, S.A, Liu, Y.C, Denholm, J, Richards, M, Rimmelzwaan, G.F, Doherty, P.C, Turner, S.J, Rossjohn, J, Kedzierska, K. | | Deposit date: | 2010-01-27 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cross-reactive CD8+ T-cell immunity between the pandemic H1N1-2009 and H1N1-1918 influenza A viruses.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3U6G
 
 | |
3U64
 
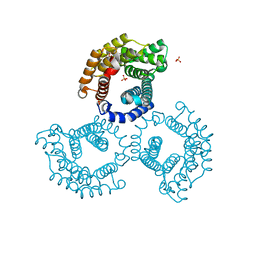 | | The Crystal Structure of Tat-T (Tp0956) | | Descriptor: | Protein TP_0956, SULFATE ION | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural, Bioinformatic, and In Vivo Analyses of Two Treponema pallidum Lipoproteins Reveal a Unique TRAP Transporter.
J.Mol.Biol., 416, 2012
|
|
3LL8
 
 | |
3U7H
 
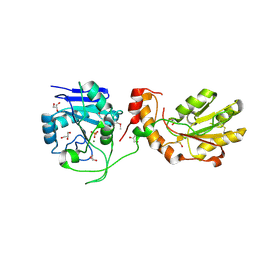 | | Crystal structure of mPNKP catalytic fragment (D170A) bound to single-stranded DNA (TCCTTp) | | Descriptor: | Bifunctional polynucleotide phosphatase/kinase, DNA, GLYCEROL, ... | | Authors: | Coquelle, N, Havali, Z, Bernstein, N, Green, R, Glover, J.N.M. | | Deposit date: | 2011-10-13 | | Release date: | 2011-12-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the phosphatase activity of polynucleotide kinase/phosphatase on single- and double-stranded DNA substrates.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3LLY
 
 | | Crystal Structure Analysis of Maclura pomifera agglutinin | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-2 chain | | Authors: | Huang, J, Xu, Z, Wang, D, Ogato, C, Hirama, T, Palczewski, K, Hazen, S.L, Lee, X, Young, N.M. | | Deposit date: | 2010-01-29 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterization of the secondary binding sites of Maclura pomifera agglutinin by glycan array and crystallographic analyses.
Glycobiology, 20, 2010
|
|
3LG8
 
 | | Crystal structure of the C-terminal part of subunit E (E101-206) from Methanocaldococcus jannaschii of A1AO ATP synthase | | Descriptor: | A-type ATP synthase subunit E | | Authors: | Balakrishna, A.M, Manimekalai, M.S.S, Hunke, C, Gayen, S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2010-01-19 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal and solution structure of the C-terminal part of the Methanocaldococcus jannaschii A1AO ATP synthase subunit E revealed by X-ray diffraction and small-angle X-ray scattering
J.Bioenerg.Biomembr., 42, 2010
|
|
3LGI
 
 | |
3U8H
 
 | | Functionally selective inhibition of Group IIA phospholipase A2 reveals a role for vimentin in regulating arachidonic acid metabolism | | Descriptor: | (S)-5-(4-BENZYLOXY-PHENYL)-4-(7-PHENYL-HEPTANOYLAMINO)-PENTANOIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Lee, L.K, Bryant, K.J, Bouveret, R, Lei, P.-W, Duff, A.P, Harrop, S.J, Huang, E.P, Harvey, R.P, Gelb, M.H, Gray, P.P, Curmi, P.M, Cunningham, A.M, Church, W.B, Scott, K.F. | | Deposit date: | 2011-10-17 | | Release date: | 2012-10-17 | | Last modified: | 2013-06-12 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Selective Inhibition of Human Group IIA-secreted Phospholipase A2 (hGIIA) Signaling Reveals Arachidonic Acid Metabolism Is Associated with Colocalization of hGIIA to Vimentin in Rheumatoid Synoviocytes.
J.Biol.Chem., 288, 2013
|
|
3LHO
 
 | |
3LI9
 
 | |
3LMT
 
 | | Crystal structure of DTD from Plasmodium falciparum | | Descriptor: | D-tyrosyl-tRNA(Tyr) deacylase, IODIDE ION | | Authors: | Manickam, Y, Bhatt, T.K, Khan, S, Sharma, A. | | Deposit date: | 2010-02-01 | | Release date: | 2010-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of D-tyrosyl-tRNATyr deacylase using home-source Cu Kalpha and moderate-quality iodide-SAD data: structural polymorphism and HEPES-bound enzyme states
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3LIS
 
 | |
3LN6
 
 | |
3TT6
 
 | |
