1TRU
 
 | |
1TXM
 
 | | SCORPION TOXIN (MAUROTOXIN) FROM SCORPIO MAURUS, NMR, 35 STRUCTURES | | Descriptor: | MAUROTOXIN | | Authors: | Blanc, E, Sabatier, J.-M, Kharrat, R, Meunier, S, El Ayeb, M, Van Rietschoten, J, Darbon, H. | | Deposit date: | 1996-11-19 | | Release date: | 1997-06-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of maurotoxin, a scorpion toxin from Scorpio maurus, with high affinity for voltage-gated potassium channels.
Proteins, 29, 1997
|
|
8BFZ
 
 | | Amyloid-beta 42 filaments extracted from the human brain with Arctic mutation (E22G) of Alzheimer's disease | ABeta42 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Yang, Y, Zhang, W.J, Murzin, A.G, Schweighauser, M, Huang, M, Lovestam, S.K.A, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Ghetti, B, Graff, C, Kumar, A, Nordberg, A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2022-10-27 | | Release date: | 2023-01-18 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of amyloid-beta filaments with the Arctic mutation (E22G) from human and mouse brains.
Acta Neuropathol, 145, 2023
|
|
1TRF
 
 | |
8BLM
 
 | | Structure of RutB | | Descriptor: | Ureidoacrylate amidohydrolase RutB | | Authors: | Rajendran, C. | | Deposit date: | 2022-11-09 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of the Ureidoacrylate Amidohydrolase RutB from Escherichia coli .
Biochemistry, 62, 2023
|
|
8AU6
 
 | |
1TNP
 
 | |
8BCQ
 
 | | N-terminal domain of Plasmodium berghei glutamyl-tRNA synthetase (native crystal structure) | | Descriptor: | GLYCEROL, Glutamate--tRNA ligase, SULFATE ION | | Authors: | Benas, P, Jaramillo Ponce, J.R, Frugier, M, Sauter, C. | | Deposit date: | 2022-10-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Solution X-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci., 32, 2023
|
|
8BLL
 
 | | Structure of RutB | | Descriptor: | ACETATE ION, Ureidoacrylate amidohydrolase RutB | | Authors: | Rajendran, C. | | Deposit date: | 2022-11-09 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural and Functional Characterization of the Ureidoacrylate Amidohydrolase RutB from Escherichia coli .
Biochemistry, 62, 2023
|
|
8BHD
 
 | | N-terminal domain of Plasmodium berghei glutamyl-tRNA synthetase (Tbxo4 derivative crystal structure) | | Descriptor: | GLYCEROL, Glutamate--tRNA ligase, SULFATE ION, ... | | Authors: | Benas, P, Jaramillo Ponce, J.R, Legrand, P, Frugier, M, Sauter, C. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Solution X-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci., 32, 2023
|
|
8BA4
 
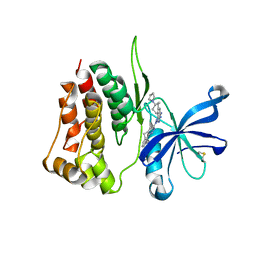 | | Crystal structure of JAK2 JH2-V617F in complex with Bemcentinib | | Descriptor: | 1-(3,4-diazatricyclo[9.4.0.0^{2,7}]pentadeca-1(11),2(7),3,5,12,14-hexaen-5-yl)-~{N}3-[(7~{S})-7-pyrrolidin-1-yl-6,7,8,9-tetrahydro-5~{H}-benzo[7]annulen-3-yl]-1,2,4-triazole-3,5-diamine, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8ATD
 
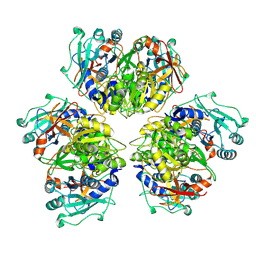 | | Wild type hexamer oxalyl-CoA synthetase (OCS) | | Descriptor: | Oxalate--CoA ligase | | Authors: | Lill, P, Burgi, J, Raunser, S, Wilmanns, M, Gatsogiannis, C. | | Deposit date: | 2022-08-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Asymmetric horseshoe-like assembly of peroxisomal yeast oxalyl-CoA synthetase.
Biol.Chem., 404, 2023
|
|
8BA3
 
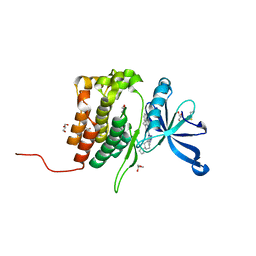 | | Crystal structure of JAK2 JH2 in complex with Bemcentinib | | Descriptor: | 1-(3,4-diazatricyclo[9.4.0.0^{2,7}]pentadeca-1(11),2(7),3,5,12,14-hexaen-5-yl)-~{N}3-[(7~{S})-7-pyrrolidin-1-yl-6,7,8,9-tetrahydro-5~{H}-benzo[7]annulen-3-yl]-1,2,4-triazole-3,5-diamine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8B64
 
 | | Cryo-EM structure of RC-LH1-PufX photosynthetic core complex from Rba. capsulatus | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Bracun, L, Yamagata, A, Shirouzu, M, Liu, L.N. | | Deposit date: | 2022-09-26 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.589 Å) | | Cite: | Cryo-EM structure of a monomeric RC-LH1-PufX supercomplex with high-carotenoid content from Rhodobacter capsulatus.
Structure, 31, 2023
|
|
8B14
 
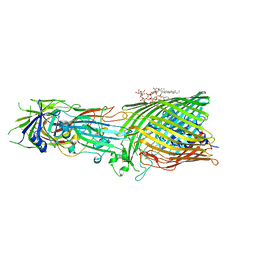 | | T5 Receptor Binding Protein pb5 in complex with its E. coli receptor FhuA | | Descriptor: | DECYLAMINE-N,N-DIMETHYL-N-OXIDE, FhuA iron-ferrichrome transporter, [(2R,3S,4R,5R,6R)-2-[[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-4-[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-2-carboxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-2-carboxy-5-oxidanyl-oxan-2-yl]oxymethyl]-5-[[(3R)-3-dodecanoyloxytetradecanoyl]amino]-4-(3-nonanoyloxypropanoyloxy)-6-[[(2R,3S,4R,5R,6R)-3-oxidanyl-4-[(3S)-3-oxidanyltetradecanoyl]oxy-5-[[(3R)-3-oxidanyltridecanoyl]amino]-6-phosphonatooxy-oxan-2-yl]methoxy]oxan-3-yl] phosphate, ... | | Authors: | Degroux, S, Effantin, G, Linares, R, Schoehn, G, Breyton, C. | | Deposit date: | 2022-09-09 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Deciphering Bacteriophage T5 Host Recognition Mechanism and Infection Trigger.
J.Virol., 97, 2023
|
|
8B2C
 
 | | Crystal structure of type I dehydroquinase from Salmonella typhi inhibited by an epoxide derivative | | Descriptor: | (1~{S},2~{R},4~{R},5~{S},6~{S})-2,4,5-trihydroxy-7-oxabicyclo[4.1.0]heptane-2-carboxylic acid, 3-dehydroquinate dehydratase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Otero, J.M, Rodriguez, A, Maneiro, M, Lence, E, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2022-09-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Quinate-based ligands for irreversible inactivation of the bacterial virulence factor DHQ1 enzyme-A molecular insight.
Front Mol Biosci, 10, 2023
|
|
8B2A
 
 | | Crystal structure of type I dehydroquinase from Staphylococcus aureus inhibited by a hydroxylamine derivative | | Descriptor: | (4R,5R)-3-amino-4,5-dihydroxy-cyclohexene-1-carboxylic acid, 3-dehydroquinate dehydratase, CHLORIDE ION, ... | | Authors: | Otero, J.M, Rodriguez, A, Maneiro, M, Lence, E, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2022-09-13 | | Release date: | 2023-02-15 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Quinate-based ligands for irreversible inactivation of the bacterial virulence factor DHQ1 enzyme-A molecular insight.
Front Mol Biosci, 10, 2023
|
|
8B2B
 
 | | Crystal structure of type I dehydroquinase from Salmonella typhi inhibited by a hydroxylamine derivative | | Descriptor: | (4R,5R)-3-amino-4,5-dihydroxy-cyclohexene-1-carboxylic acid, 3-dehydroquinate dehydratase, SODIUM ION | | Authors: | Otero, J.M, Rodriguez, A, Maneiro, M, Lence, E, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2022-09-13 | | Release date: | 2023-02-15 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Quinate-based ligands for irreversible inactivation of the bacterial virulence factor DHQ1 enzyme-A molecular insight.
Front Mol Biosci, 10, 2023
|
|
8ATX
 
 | |
8B8U
 
 | |
8BFY
 
 | | ABC transporter binding protein CebE from Streptomyces scabiei in complex with cellotriose | | Descriptor: | CITRIC ACID, GLYCEROL, Putative secreted cellobiose-binding (Transport system associated), ... | | Authors: | Rigali, S, Jourdan, S, Kerff, F. | | Deposit date: | 2022-10-27 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Common scab disease: structural basis of elicitor recognition in pathogenic Streptomyces species.
Microbiol Spectr, 11, 2023
|
|
8BBU
 
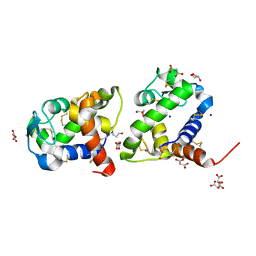 | | Crystal structure of medical leech destabilase (high salt) | | Descriptor: | GLYCEROL, Lysozyme, MALONATE ION, ... | | Authors: | Marin, E, Bukhdruker, S, Manuvera, V, Kornilov, D, Zinovev, E, Bobrovsky, P, Lazarev, V, Borshchevskiy, V. | | Deposit date: | 2022-10-14 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural insights into thrombolytic activity of destabilase from medicinal leech.
Sci Rep, 13, 2023
|
|
8AUD
 
 | |
8B5Y
 
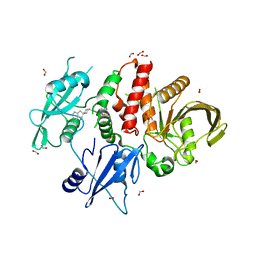 | | SHP2 in complex with allosteric imidazopyrazine inhibitor | | Descriptor: | (1~{S})-1'-[5-[2-(trifluoromethyl)pyridin-3-yl]sulfanyl-3~{H}-imidazo[4,5-b]pyrazin-2-yl]spiro[1,3-dihydroindene-2,4'-piperidine]-1-amine, FORMIC ACID, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Torrente, E, Petrocchi, A. | | Deposit date: | 2022-09-25 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of a Novel Series of Imidazopyrazine Derivatives as Potent SHP2 Allosteric Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8B4N
 
 | |
