8VQW
 
 | |
8OZ9
 
 | |
5DVA
 
 | |
8SSC
 
 | |
8P2N
 
 | |
7W8B
 
 | |
8VFU
 
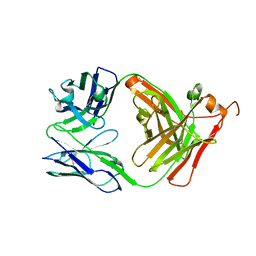 | |
7ZCD
 
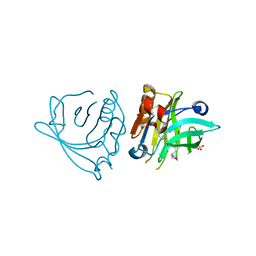 | |
6MV7
 
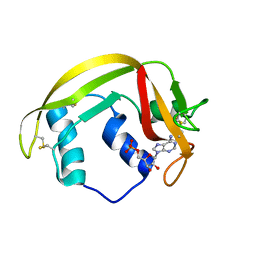 | | Crystal structure of RNAse 6 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ribonuclease K6 | | Authors: | Couture, J.-F, Doucet, N. | | Deposit date: | 2018-10-24 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Insights into Structural and Dynamical Changes Experienced by Human RNase 6 upon Ligand Binding.
Biochemistry, 59, 2020
|
|
7MWU
 
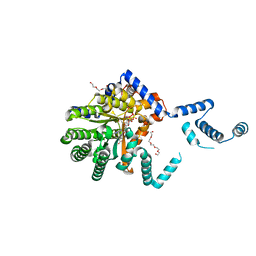 | |
5V2I
 
 | |
7ZE9
 
 | | Structure of an AA16 LPMO-like protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COPPER (II) ION, ... | | Authors: | Huang, Z, Banerjee, S, Muderspach, S.J, Sun, P, van Berkel, W.J.H, Kabel, M.A, Lo Leggio, L. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.646 Å) | | Cite: | AA16 Oxidoreductases Boost Cellulose-Active AA9 Lytic Polysaccharide Monooxygenases from Myceliophthora thermophila.
Acs Catalysis, 13, 2023
|
|
7NDX
 
 | | Crystal structure of the human HSP40 DNAJB1-CTDs in complex with a peptide of NudC | | Descriptor: | 1,2-ETHANEDIOL, DnaJ homolog subfamily B member 1, Nuclear migration protein nudC | | Authors: | Delhommel, F, Zak, K.M, Popowicz, G.M, Sattler, M. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | NudC guides client transfer between the Hsp40/70 and Hsp90 chaperone systems.
Mol.Cell, 82, 2022
|
|
8SSG
 
 | |
5V2Z
 
 | | Ethylene forming enzyme in complex with manganese, 2-oxoadipic acid and L-arginine | | Descriptor: | 2-OXOADIPIC ACID, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ARGININE, ... | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
6MVU
 
 | |
7N14
 
 | | Crystal structure of 4-(1H-1,2,4-triazol-1-yl)benzoic acid-bound CYP199A4 | | Descriptor: | 4-(1H-1,2,4-triazol-1-yl)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2021-05-26 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.537 Å) | | Cite: | To Be, or Not to Be, an Inhibitor: A Comparison of Azole Interactions with and Oxidation by a Cytochrome P450 Enzyme.
Inorg.Chem., 61, 2022
|
|
8W0C
 
 | |
8W0D
 
 | |
7MU8
 
 | | Structure of the minimally glycosylated human CEACAM1 N-terminal domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carcinoembryonic antigen-related cell adhesion molecule 1, GLYCEROL, ... | | Authors: | Belcher Dufrisne, M, Swope, N, Kieber, M, Yang, J.Y, Han, J, Li, J, Moremen, K.W, Prestegard, J.H, Columbus, L. | | Deposit date: | 2021-05-14 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Human CEACAM1 N-domain dimerization is independent from glycan modifications.
Structure, 30, 2022
|
|
5DXA
 
 | |
8SWT
 
 | |
6N6Q
 
 | |
7PK8
 
 | | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria | | Descriptor: | (2S,4S)-4-fluoranyl-N-[(3R,5R)-5-[[[4-[2-(4-methylphenyl)ethynyl]phenyl]carbonylamino]methyl]-1-(2-methylpropanoyl)pyrrolidin-3-yl]pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Ryan, M.D, Pallin, T.D, Lamers, M.B.A.C, Leonard, P.M. | | Deposit date: | 2021-08-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria
To Be Published
|
|
7MZV
 
 | | Structure of yeast pseudouridine synthase 7 (PUS7) | | Descriptor: | Multisubstrate pseudouridine synthase 7, SULFATE ION | | Authors: | Purchal, M, Koutmos, M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Pseudouridine synthase 7 is an opportunistic enzyme that binds and modifies substrates with diverse sequences and structures.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
