1ISZ
 
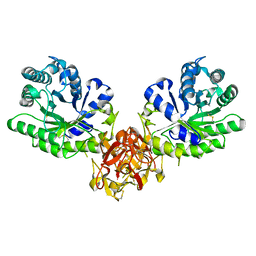 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with galactose | | Descriptor: | beta-D-galactopyranose, endo-1,4-beta-D-xylanase | | Authors: | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2001-12-27 | | Release date: | 2002-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
1IDC
 
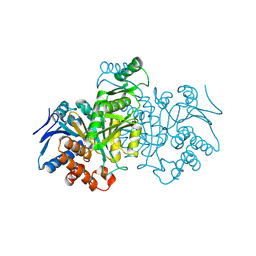 | | ISOCITRATE DEHYDROGENASE FROM E.COLI (MUTANT K230M), STEADY-STATE INTERMEDIATE COMPLEX DETERMINED BY LAUE CRYSTALLOGRAPHY | | Descriptor: | 2-OXALOSUCCINIC ACID, ISOCITRATE DEHYDROGENASE, MAGNESIUM ION | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
1IEL
 
 | | Crystal Structure of AmpC beta-lactamase from E. coli in Complex with Ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, PHOSPHATE ION, beta-lactamase | | Authors: | Powers, R.A, Caselli, E, Focia, P.J, Prati, F, Shoichet, B.K. | | Deposit date: | 2001-04-10 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of ceftazidime and its transition-state analogue in complex with AmpC beta-lactamase: implications for resistance mutations and inhibitor design.
Biochemistry, 40, 2001
|
|
1ISX
 
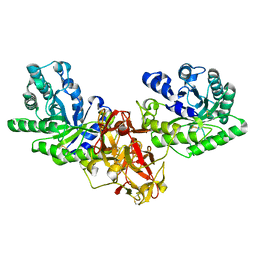 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylotriose | | Descriptor: | beta-D-xylopyranose, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2001-12-27 | | Release date: | 2002-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
2RH2
 
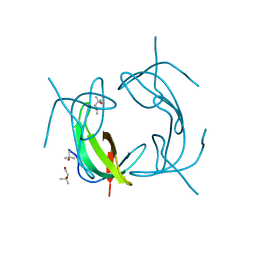 | | High Resolution DHFR R-67 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Dihydrofolate reductase type 2 | | Authors: | Krahn, J.M, London, R.E. | | Deposit date: | 2007-10-05 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Crystal Structure of a Type II Dihydrofolate Reductase Catalytic Ternary Complex
Biochemistry, 46, 2007
|
|
1IEM
 
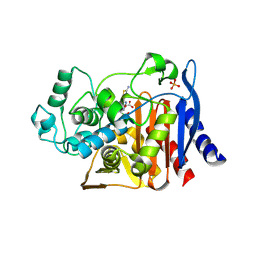 | | Crystal Structure of AmpC beta-lactamase from E. coli in Complex with a Boronic Acid Inhibitor (1, CefB4) | | Descriptor: | PHOSPHATE ION, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE, beta-lactamase | | Authors: | Powers, R.A, Caselli, E, Focia, P.J, Prati, F, Shoichet, B.K. | | Deposit date: | 2001-04-10 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of ceftazidime and its transition-state analogue in complex with AmpC beta-lactamase: implications for resistance mutations and inhibitor design.
Biochemistry, 40, 2001
|
|
2RK1
 
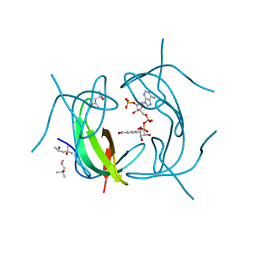 | | DHFR R67 Complexed with NADP and dihydrofolate | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, DIHYDROFOLIC ACID, Dihydrofolate reductase type 2, ... | | Authors: | Krahn, J.M, London, R.E. | | Deposit date: | 2007-10-16 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structure of a type II dihydrofolate reductase catalytic ternary complex.
Biochemistry, 46, 2007
|
|
2RK2
 
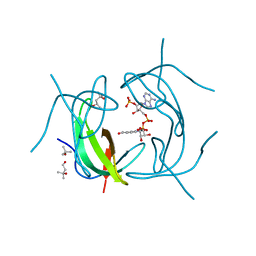 | | DHFR R-67 complexed with NADP | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Dihydrofolate reductase type 2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Krahn, J.M, London, R.E. | | Deposit date: | 2007-10-16 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a type II dihydrofolate reductase catalytic ternary complex.
Biochemistry, 46, 2007
|
|
1J54
 
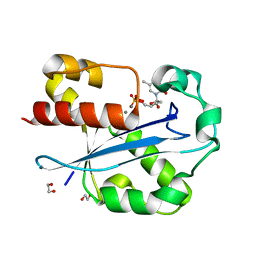 | | Structure of the N-terminal exonuclease domain of the epsilon subunit of E.coli DNA polymerase III at pH 5.8 | | Descriptor: | 1,2-ETHANEDIOL, DNA polymerase III, epsilon chain, ... | | Authors: | Hamdan, S, Carr, P.D, Brown, S.E, Ollis, D.L, Dixon, N.E. | | Deposit date: | 2002-01-22 | | Release date: | 2002-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Proofreading during Replication of the Escherichia coli Chromosome
Structure, 10, 2002
|
|
1ISY
 
 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with glucose | | Descriptor: | beta-D-glucopyranose, endo-1,4-beta-D-xylanase | | Authors: | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2001-12-27 | | Release date: | 2002-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
1IGP
 
 | |
1JDZ
 
 | | CRYSTAL STRUCTURE OF 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE WITH FORMYCIN B AND SULFATE ION | | Descriptor: | 5'-METHYLTHIOADENOSINE PHOSPHORYLASE, FORMYCIN B, SULFATE ION | | Authors: | Appleby, T.C, Mathews, I.I, Porcelli, M, Cacciapuoti, G, Ealick, S.E. | | Deposit date: | 2001-06-15 | | Release date: | 2001-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of a hyperthermophilic 5'-deoxy-5'-methylthioadenosine phosphorylase from Sulfolobus solfataricus.
J.Biol.Chem., 276, 2001
|
|
1JE0
 
 | | CRYSTAL STRUCTURE OF 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE COMPLEXED WITH PHOSPHATE AND TRIS MOLECULE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-METHYLTHIOADENOSINE PHOSPHORYLASE, PHOSPHATE ION | | Authors: | Appleby, T.C, Mathews, I.I, Porcelli, M, Cacciapuoti, G, Ealick, S.E. | | Deposit date: | 2001-06-15 | | Release date: | 2001-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three-dimensional structure of a hyperthermophilic 5'-deoxy-5'-methylthioadenosine phosphorylase from Sulfolobus solfataricus.
J.Biol.Chem., 276, 2001
|
|
2BH7
 
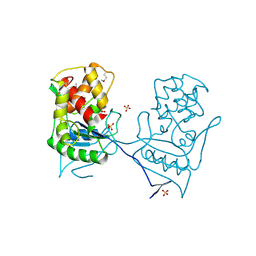 | | Crystal structure of a SeMet derivative of AmiD at 2.2 angstroms | | Descriptor: | N-ACETYLMURAMOYL-L-ALANINE AMIDASE, SULFATE ION, ZINC ION | | Authors: | Petrella, S, Herman, R, Sauvage, E, Genereux, C, Pennartz, A, Joris, B, Charlier, P. | | Deposit date: | 2005-01-07 | | Release date: | 2006-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Specific Structural Features of the N-Acetylmuramoyl-L-Alanine Amidase Amid from Escherichia Coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
1IX9
 
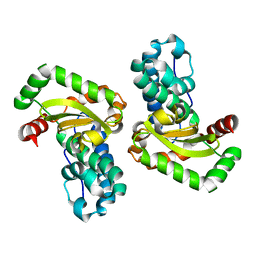 | | Crystal Structure of the E. coli Manganase(III) superoxide dismutase mutant Y174F at 0.90 angstroms resolution. | | Descriptor: | MANGANESE (II) ION, Superoxide Dismutase | | Authors: | Anderson, B.F, Edwards, R.A, Whittaker, M.M, Whittaker, J.W, Baker, E.N, Jameson, G.B. | | Deposit date: | 2002-06-17 | | Release date: | 2002-12-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structures at 0.90 A resolution of the oxidised and reduced forms of the Y174F mutant of the manganese superoxide dismutase from Escherichia coli
To be Published
|
|
1IXB
 
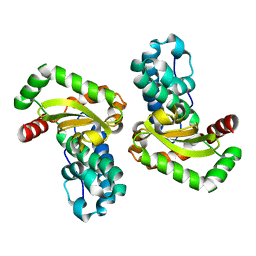 | | CRYSTAL STRUCTURE OF THE E. COLI MANGANESE(II) SUPEROXIDE DISMUTASE MUTANT Y174F AT 0.90 ANGSTROMS RESOLUTION. | | Descriptor: | MANGANESE ION, 1 HYDROXYL COORDINATED, SUPEROXIDE DISMUTASE | | Authors: | Anderson, B.F, Edwards, R.A, Whittaker, M.M, Whittaker, J.W, Baker, E.N, Jameson, G.B. | | Deposit date: | 2002-06-18 | | Release date: | 2002-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structures at 0.90 A resolution of the oxidised and reduced forms of the Y174F mutant of the manganese superoxide dismutase from Escherichia coli
To be Published
|
|
2SCU
 
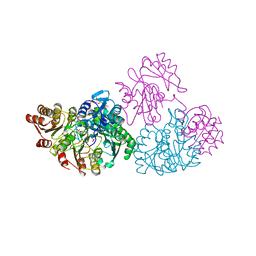 | | A detailed description of the structure of Succinyl-COA synthetase from Escherichia coli | | Descriptor: | COENZYME A, PROTEIN (SUCCINYL-COA LIGASE), SULFATE ION | | Authors: | Fraser, M.E, Wolodko, W.T, James, M.N.G, Bridger, W.A. | | Deposit date: | 1998-09-24 | | Release date: | 1999-08-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A detailed structural description of Escherichia coli succinyl-CoA synthetase.
J.Mol.Biol., 285, 1999
|
|
4CAU
 
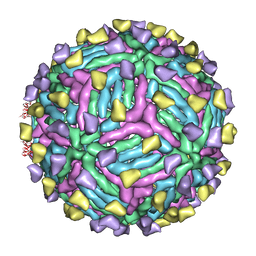 | | THREE-DIMENSIONAL STRUCTURE OF DENGUE VIRUS SEROTYPE 1 COMPLEXED WITH 2 HMAB 14C10 FAB | | Descriptor: | ENVELOPE PROTEIN E, FAB 14C10 | | Authors: | Teoh, E.P, Kukkaro, P, Teo, E.W, Lim, A.P, Tan, T.T, Yip, A, Schul, W, Aung, M, Kostyuchenko, V.A, Leo, Y.S, Chan, S.H, Smith, K.G, Chan, A.H, Zou, G, Ooi, E.E, Kemeny, D.M, Tan, G.K, Ng, J.K, Ng, M.L, Alonso, S, Fisher, D, Shi, P.Y, Hanson, B.J, Lok, S.M, Macary, P.A. | | Deposit date: | 2013-10-09 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | The Structural Basis for Serotype-Specific Neutralization of Dengue Virus by a Human Antibody.
Sci.Trans.Med, 4, 2012
|
|
1JBZ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A DUAL-WAVELENGTH EMISSION GREEN FLUORESCENT PROTEIN VARIANT AT HIGH PH | | Descriptor: | 1,2-ETHANEDIOL, GREEN FLUORESCENT PROTEIN, MAGNESIUM ION | | Authors: | Hanson, G.T, McAnaney, T.B, Park, E.S, Rendell, M.E.P, Yarbrough, D.K, Chu, S, Xi, L, Boxer, S.G, Montrose, M.H, Remington, S.J. | | Deposit date: | 2001-06-07 | | Release date: | 2003-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Green Fluorescent Protein Variants as Ratiometric Dual Emission pH Sensors. 1. Structural Characterization and Preliminary Application.
Biochemistry, 41, 2002
|
|
7AJF
 
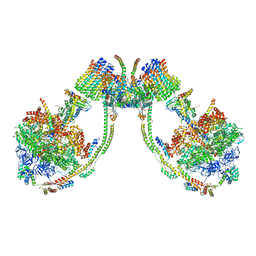 | | bovine ATP synthase dimer state2:state2 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ATP synthase F(0) complex subunit B1, mitochondrial, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-03 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (8.45 Å) | | Cite: | Interface mobility between monomers in dimeric bovine ATP synthase participates in the ultrastructure of inner mitochondrial membranes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2RG1
 
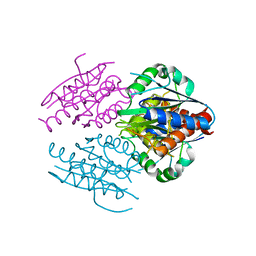 | | Crystal structure of E. coli WrbA apoprotein | | Descriptor: | CHLORIDE ION, Flavoprotein WrbA | | Authors: | Kuta Smatanova, I, Wolfova, J, Brynda, J, Lapkouski, M, Mesters, J.R, Grandori, R, Carey, J. | | Deposit date: | 2007-10-02 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural organization of WrbA in apo- and holoprotein crystals.
Biochim.Biophys.Acta, 1794, 2009
|
|
2BS0
 
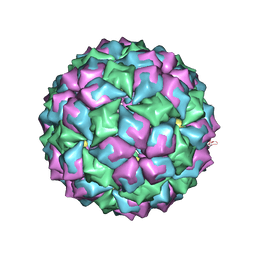 | | MS2 (N87AE89K mutant) - Variant Qbeta RNA hairpin complex | | Descriptor: | 5'-R(*AP*UP*GP*CP*AP*UP*GP*UP*CP*UP *AP*AP*GP*AP*CP*UP*GP*CP*AP*U)-3', COAT PROTEIN | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-05-13 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Basis of RNA Binding Discrimination between Bacteriophages Qbeta and MS2.
Structure, 14, 2006
|
|
1INJ
 
 | | CRYSTAL STRUCTURE OF THE APO FORM OF 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL (CDP-ME) SYNTHETASE (YGBP) INVOLVED IN MEVALONATE INDEPENDENT ISOPRENOID BIOSYNTHESIS | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL SYNTHETASE, CALCIUM ION | | Authors: | Richard, S.B, Bowman, M.E, Kwiatkowski, W, Kang, I, Chow, C, Lillo, A, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-05-14 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of 4-diphosphocytidyl-2-C- methylerythritol synthetase involved in mevalonate- independent isoprenoid biosynthesis.
Nat.Struct.Biol., 8, 2001
|
|
2BS1
 
 | | MS2 (N87AE89K mutant) - Qbeta RNA hairpin complex | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP *UP*AP*CP*CP*CP*AP*UP*GP*U)-3', MS2 COAT PROTEIN | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-05-13 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of RNA Binding Discrimination between Bacteriophages Qbeta and MS2.
Structure, 14, 2006
|
|
2BNY
 
 | | MS2 (N87A mutant) - RNA hairpin complex | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP *UP*AP*CP*CP*CP*AP*UP*GP*U)-3', MS2 COAT PROTEIN | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-04-06 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of RNA Binding Discrimination between Bacteriophages Qbeta and MS2.
Structure, 14, 2006
|
|
