4NHZ
 
 | | Crystal structure of glutathione transferase BBTA-3750 from Bradyrhizobium sp., Target EFI-507290, with one glutathione bound | | Descriptor: | GLUTATHIONE, Putative glutathione S-transferase enzyme with thioredoxin-like domain | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-05 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Crystal Structure of Glutathione Transferase Bbta-3750 from Bradyrhizobium Sp., Target Efi-507290
To be Published
|
|
3VK9
 
 | | Crystal structure of delta-class glutathione transferase from silkmoth | | Descriptor: | GLYCEROL, Glutathione S-transferase delta | | Authors: | Kakuta, Y, Usuda, K, Higashiura, A, Suzuki, M, Nakagawa, A, Kimura, M, Yamamoto, K. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for catalytic activity of a silkworm Delta-class glutathione transferase
Biochim.Biophys.Acta, 1820, 2012
|
|
4O92
 
 | | Crystal structure of a Glutathione S-transferase from Pichia kudriavzevii (Issatchenkia orientalis), target EFI-501747 | | Descriptor: | Glutathione S-transferase, SULFATE ION | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Attonito, J.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-31 | | Release date: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of a Glutathione S-transferase from Pichia kudriavzevii (Issatchenkia orientalis), target EFI-501747
TO BE PUBLISHED
|
|
3VWX
 
 | | Structural analysis of an epsilon-class glutathione S-transferase from housefly, Musca domestica | | Descriptor: | GLUTATHIONE, Glutathione s-transferase 6B | | Authors: | Nakamura, C, Sue, M, Miyamoto, T, Yajima, S. | | Deposit date: | 2012-09-05 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of an epsilon-class glutathione transferase from housefly, Musca domestica
Biochem.Biophys.Res.Commun., 430, 2013
|
|
4PNF
 
 | | Glutathione S-Transferase from Drosophila melanogaster - isozyme E6 | | Descriptor: | GLUTATHIONE, RE21095p | | Authors: | Scian, M, Le Trong, I, Mannervik, B, Atkins, W.M, Stenkamp, R.E. | | Deposit date: | 2014-05-23 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Comparison of epsilon- and delta-class glutathione S-transferases: the crystal structures of the glutathione S-transferases DmGSTE6 and DmGSTE7 from Drosophila melanogaster.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4PNG
 
 | | Glutathione S-transferase from Drosophila melanogaster - isozyme E7 | | Descriptor: | L-GAMMA-GLUTAMYL-3-SULFINO-L-ALANYLGLYCINE, LD04004p | | Authors: | Scian, M, Le Trong, I, Mannervik, B, Atkins, W.M, Stenkamp, R.E. | | Deposit date: | 2014-05-23 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Comparison of epsilon- and delta-class glutathione S-transferases: the crystal structures of the glutathione S-transferases DmGSTE6 and DmGSTE7 from Drosophila melanogaster.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WYW
 
 | | Structural characterization of catalytic site of a Nilaparvata lugens delta-class glutathione transferase | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase | | Authors: | Yamamoto, K, Higashiura, A, Nakagawa, A. | | Deposit date: | 2014-09-09 | | Release date: | 2015-01-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of the catalytic site of a Nilaparvata lugens delta-class glutathione transferase.
Arch.Biochem.Biophys., 566C, 2014
|
|
3ZFL
 
 | | Crystal structure of the V58A mutant of human class alpha glutathione transferase in the apo form | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1 | | Authors: | Parbhoo, N, Fanucchi, S, Achilonu, I.A, Fernandes, M.A, Gildenhuys, S, Dirr, H.W. | | Deposit date: | 2012-12-12 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of the V58A Mutant of Human Class Alpha Glutathione Transferase in the Apo Form
To be Published
|
|
3ZML
 
 | |
3ZMK
 
 | |
3ZFB
 
 | | Crystal structure of the I75A mutant of human class alpha glutathione transferase in the apo form | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1 | | Authors: | Parbhoo, N, Fanucchi, S, Achilonu, I.A, Fernandes, M.A, Gildenhuys, S, Dirr, H.W. | | Deposit date: | 2012-12-11 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of the I75A Mutant of Human Class Alpha Glutathione Transferase in the Apo Form
To be Published
|
|
4QQ7
 
 | |
4ACS
 
 | | Crystal structure of mutant GST A2-2 with enhanced catalytic efficiency with azathioprine | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE A2 | | Authors: | Zhang, W, Moden, O, Tars, K, Mannervik, B. | | Deposit date: | 2011-12-19 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based redesign of GST A2-2 for enhanced catalytic efficiency with azathioprine.
Chem.Biol., 19, 2012
|
|
4RI7
 
 | | Crystal structure of poplar glutathione transferase F1 mutant SER 13 CYS | | Descriptor: | GLUTATHIONE, Phi class glutathione transferase GSTF1 | | Authors: | Pegeot, H, Mathiot, S, Didierjean, C, Rouhier, N. | | Deposit date: | 2014-10-05 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | The poplar Phi class glutathione transferase: expression, activity and structure of GSTF1.
Front Plant Sci, 5, 2014
|
|
4RI6
 
 | | Crystal structure of poplar glutathione transferase F1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, Phi class glutathione transferase GSTF1 | | Authors: | Pegeot, H, Koh, C.S, Didierjean, C, Rouhier, N. | | Deposit date: | 2014-10-05 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | The poplar Phi class glutathione transferase: expression, activity and structure of GSTF1.
Front Plant Sci, 5, 2014
|
|
4BL7
 
 | |
8A0O
 
 | |
8A0I
 
 | |
8A0R
 
 | |
8A08
 
 | |
8A0P
 
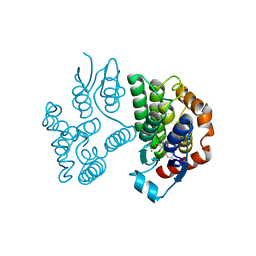 | | Crystal structure of poplar glutathione transferase U20 in complex with morin | | Descriptor: | 2-[2,4-bis(oxidanyl)phenyl]-3,5,7-tris(oxidanyl)chromen-4-one, CHLORIDE ION, Glutathione transferase | | Authors: | Didierjean, C, Favier, F. | | Deposit date: | 2022-05-30 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.686 Å) | | Cite: | Biochemical and Structural Insights on the Poplar Tau Glutathione Transferase GSTU19 and 20 Paralogs Binding Flavonoids.
Front Mol Biosci, 9, 2022
|
|
8A0Q
 
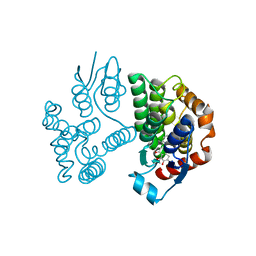 | |
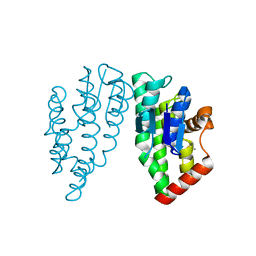
7ZZN
 
 | |
8ALS
 
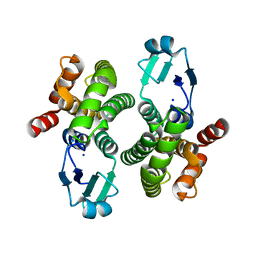 | |
8AGQ
 
 | | Crystal structure of anthocyanin-related GSTF8 from Populus trichocarpa in complex with (-)-catechin and glutathione | | Descriptor: | (2~{S},3~{R})-2-[3,4-bis(oxidanyl)phenyl]-3,4-dihydro-2~{H}-chromene-3,5,7-triol, GLUTATHIONE, Glutathione transferase, ... | | Authors: | Eichenberger, M, Hueppi, S, Schwander, T, Mittl, P, Buller, M.R. | | Deposit date: | 2022-07-20 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.093 Å) | | Cite: | The catalytic role of glutathione transferases in heterologous anthocyanin biosynthesis.
Nat Catal, 6, 2023
|
|
