1DVS
 
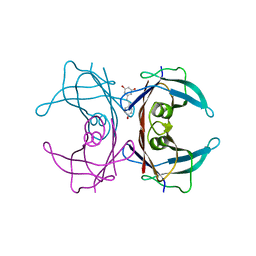 | | CRYSTAL STRUCTURE OF HUMAN TRANSTHYRETIN IN COMPLEX WITH RESVERATROL | | Descriptor: | RESVERATROL, TRANSTHYRETIN | | Authors: | Klabunde, T, Petrassi, H.M, Oza, V.B, Kelly, J.W, Sacchettini, J.C. | | Deposit date: | 2000-01-22 | | Release date: | 2001-01-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational design of potent human transthyretin amyloid disease inhibitors.
Nat.Struct.Biol., 7, 2000
|
|
2OVC
 
 | |
3LU1
 
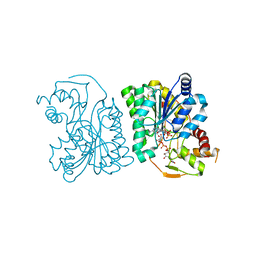 | | Crystal Structure Analysis of WbgU: a UDP-GalNAc 4-epimerase | | Descriptor: | GLYCINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Bhatt, V.S, Guo, C.Y, Zhao, G, Yi, W, Liu, Z.J, Wang, P.G. | | Deposit date: | 2010-02-16 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Altered architecture of substrate binding region defines the unique specificity of UDP-GalNAc 4-epimerases.
Protein Sci., 20, 2011
|
|
3VHZ
 
 | |
1OLG
 
 | |
3W0R
 
 | | Crystal structure of a thermostable mutant of aminoglycoside phosphotransferase APH(4)-Ia (N202A), ternary complex with AMP-PNP and hygromycin B | | Descriptor: | HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Iino, D, Takakura, Y, Fukano, K, Sasaki, Y, Hoshino, T, Ohsawa, K, Nakamura, A, Yajima, S. | | Deposit date: | 2012-11-02 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the ternary complex of APH(4)-Ia/Hph with hygromycin B and an ATP analog using a thermostable mutant.
J.Struct.Biol., 183, 2013
|
|
1K1E
 
 | |
1OLH
 
 | |
4QXV
 
 | | CRYSTAL STRUCTURE of HUMAN TRANSTHYRETIN IN COMPLEX WITH LUTEOLIN AT 1.1 A RESOLUTION | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, GLYCEROL, SODIUM ION, ... | | Authors: | Begum, A, Olofsson, A, Sauer-Eriksson, A.E. | | Deposit date: | 2014-07-22 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | The flavonoid luteolin, but not luteolin-7-o-glucoside, prevents a transthyretin mediated toxic response.
Plos One, 10, 2015
|
|
1PM9
 
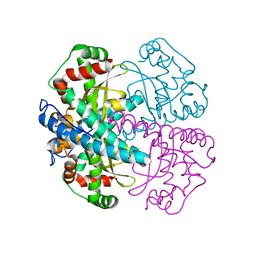 | | CRYSTAL STRUCTURE OF HUMAN MNSOD H30N, Y166F MUTANT | | Descriptor: | MANGANESE (III) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Fan, L, Tainer, J.A. | | Deposit date: | 2003-06-10 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Amino acid substitution at the dimeric interface of human manganese superoxide dismutase
J.Biol.Chem., 279, 2004
|
|
4QYA
 
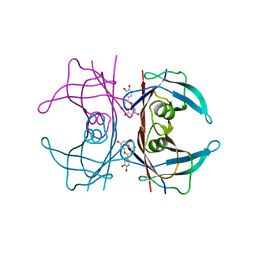 | | Crystal structure of human transthyretin variant V30M in complex with luteolin | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, SODIUM ION, Transthyretin | | Authors: | Begum, A, Olofsson, A, Sauer-Eriksson, A.E. | | Deposit date: | 2014-07-24 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The flavonoid luteolin, but not luteolin-7-o-glucoside, prevents a transthyretin mediated toxic response.
Plos One, 10, 2015
|
|
1Q0C
 
 | | Anerobic Substrate Complex of Homoprotocatechuate 2,3-Dioxygenase from Brevibacterium fuscum. (Complex with 3,4-Dihydroxyphenylacetate) | | Descriptor: | 2-(3,4-DIHYDROXYPHENYL)ACETIC ACID, FE (III) ION, homoprotocatechuate 2,3-dioxygenase | | Authors: | Vetting, M.W, Wackett, L.P, Que, L, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2003-07-15 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic comparison of manganese- and iron-dependent homoprotocatechuate 2,3-dioxygenases.
J.Bacteriol., 186, 2004
|
|
1KOL
 
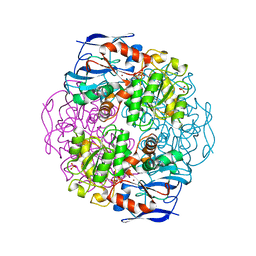 | | Crystal structure of formaldehyde dehydrogenase | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ZINC ION, ... | | Authors: | Tanaka, N, Kusakabe, Y, Ito, K, Yoshimoto, T, Nakamura, K.T. | | Deposit date: | 2001-12-21 | | Release date: | 2002-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Formaldehyde Dehydrogenase from Pseudomonas putida: the Structural Origin of the Tightly Bound Cofactor in Nicotinoprotein Dehydrogenases
J.mol.biol., 324, 2002
|
|
1COJ
 
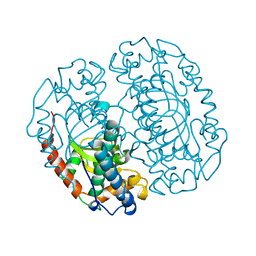 | | FE-SOD FROM AQUIFEX PYROPHILUS, A HYPERTHERMOPHILIC BACTERIUM | | Descriptor: | FE (III) ION, PROTEIN (SUPEROXIDE DISMUTASE) | | Authors: | Lim, J.H, Yu, Y.G, Kim, S.-H, Cho, S.-J, Ahn, B.Y, Han, Y.S, Cho, Y. | | Deposit date: | 1999-05-28 | | Release date: | 1999-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of an Fe-superoxide dismutase from the hyperthermophile Aquifex pyrophilus at 1.9 A resolution: structural basis for thermostability.
J.Mol.Biol., 270, 1997
|
|
1NPL
 
 | | MANNOSE-SPECIFIC AGGLUTININ (LECTIN) FROM DAFFODIL (NARCISSUS PSEUDONARCISSUS) BULBS IN COMPLEX WITH MANNOSE-ALPHA1,3-MANNOSE | | Descriptor: | PHOSPHATE ION, PROTEIN (AGGLUTININ), alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose | | Authors: | Sauerborn, M.K, Wright, L.M, Reynolds, C.D, Grossmann, J.G, Rizkallah, P.J. | | Deposit date: | 1998-12-17 | | Release date: | 1998-12-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into carbohydrate recognition by Narcissus pseudonarcissus lectin: the crystal structure at 2 A resolution in complex with alpha1-3 mannobiose.
J.Mol.Biol., 290, 1999
|
|
1CR7
 
 | | PEANUT LECTIN-LACTOSE COMPLEX MONOCLINIC FORM | | Descriptor: | CALCIUM ION, LECTIN, MANGANESE (II) ION, ... | | Authors: | Ravishankar, R, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 1999-08-14 | | Release date: | 2001-04-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the peanut lectin-lactose complex at acidic pH: retention of unusual quaternary structure, empty and carbohydrate bound combining sites, molecular mimicry and crystal packing directed by interactions at the combining site.
Proteins, 43, 2001
|
|
7R3B
 
 | | S-adenosylmethionine synthetase from Lactobacillus plantarum complexed with AMPPNP, methionine and SAM | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, MAGNESIUM ION, PHENYLALANINE, ... | | Authors: | Shahar, A, Kleiner, D, Bershtein, S, Zarivach, R. | | Deposit date: | 2022-02-07 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Evolution of homo-oligomerization of methionine S-adenosyltransferases is replete with structure-function constrains.
Protein Sci., 31, 2022
|
|
7R2W
 
 | | Mutant S-adenosylmethionine synthetase from E.coli complexed with AMPPNP and methionine | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Shahar, A, Kleiner, D, Bershtein, S, Zarivach, R. | | Deposit date: | 2022-02-06 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolution of homo-oligomerization of methionine S-adenosyltransferases is replete with structure-function constrains.
Protein Sci., 31, 2022
|
|
2EWM
 
 | |
2EW8
 
 | |
1SCU
 
 | | THE CRYSTAL STRUCTURE OF SUCCINYL-COA SYNTHETASE FROM ESCHERICHIA COLI AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | COENZYME A, SUCCINYL-COA SYNTHETASE, ALPHA SUBUNIT, ... | | Authors: | Wolodko, W.T, Fraser, M.E, James, M.N.G, Bridger, W.A. | | Deposit date: | 1993-11-18 | | Release date: | 1995-04-20 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of succinyl-CoA synthetase from Escherichia coli at 2.5-A resolution.
J.Biol.Chem., 269, 1994
|
|
8C6Z
 
 | | necrotic enteritis associated Clostridium p. chitinase F8UNI4 in complex with inhibitor bisdionin C | | Descriptor: | 1,1'-PROPANE-1,3-DIYLBIS(3,7-DIMETHYL-3,7-DIHYDRO-1H-PURINE-2,6-DIONE), CADMIUM ION, COBALT (II) ION, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Contribution and structural characterization of Chitinases to Clostridium perfringens Pathogenesis in Broilers
To Be Published
|
|
4GCL
 
 | | structure of no-dna factor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*AP*GP*TP*GP*AP*GP*TP*AP*CP*TP*CP*AP*CP*T)-3'), Nucleoid occlusion factor SlmA | | Authors: | Schumacher, M.A. | | Deposit date: | 2012-07-30 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1EFM
 
 | |
7RWS
 
 | |
