1PET
 
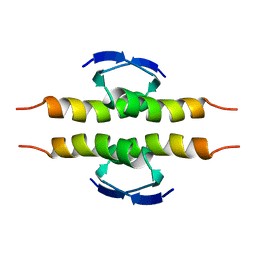 | | NMR SOLUTION STRUCTURE OF THE TETRAMERIC MINIMUM TRANSFORMING DOMAIN OF P53 | | Descriptor: | TUMOR SUPPRESSOR P53 | | Authors: | Lee, W, Harvey, T.S, Yin, Y, Yau, P, Litchfield, D, Arrowsmith, C.H. | | Deposit date: | 1994-11-24 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tetrameric minimum transforming domain of p53.
Nat.Struct.Biol., 1, 1994
|
|
4LCR
 
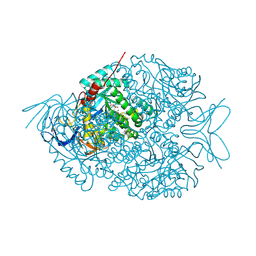 | | The crystal structure of di-Zn dihydropyrimidinase in complex with NCBA | | Descriptor: | Chromosome 8 SCAF14545, whole genome shotgun sequence, N-(AMINOCARBONYL)-BETA-ALANINE, ... | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2013-06-22 | | Release date: | 2013-09-18 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of vertebrate dihydropyrimidinase and complexes from Tetraodon nigroviridis with lysine carbamylation: metal and structural requirements for post-translational modification and function.
J.Biol.Chem., 288, 2013
|
|
3BQY
 
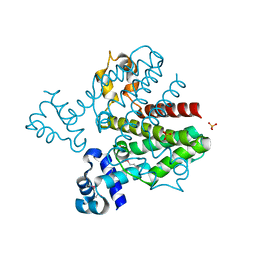 | | Crystal structure of a possible TetR family transcriptional regulator from Streptomyces coelicolor A3(2). | | Descriptor: | ACETIC ACID, PHOSPHATE ION, Putative TetR family transcriptional regulator | | Authors: | Cuff, M.E, Skarina, T, Kagan, O, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-20 | | Release date: | 2008-01-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of a possible TetR family transcriptional regulator from Streptomyces coelicolor A3(2).
TO BE PUBLISHED
|
|
1VGL
 
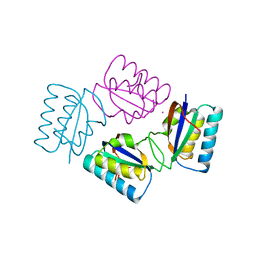 | | Crystal structure of tetrameric KaiB from T.elongatus BP-1 | | Descriptor: | Circadian clock protein kaiB, MERCURY (II) ION | | Authors: | Iwase, R, Imada, K, Hayashi, F, Uzumaki, T, Namba, K, Ishiura, M. | | Deposit date: | 2004-04-27 | | Release date: | 2005-08-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functionally important substructures of circadian clock protein KaiB in a unique tetramer complex.
J.Biol.Chem., 280, 2005
|
|
8ZBA
 
 | |
6DHH
 
 | | RT XFEL structure of Photosystem II 400 microseconds after the second illumination at 2.2 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
6DHF
 
 | | RT XFEL structure of the one-flash state of Photosystem II (1F, S2-rich) at 2.08 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
6DHO
 
 | | RT XFEL structure of the two-flash state of Photosystem II (2F, S3-rich) at 2.07 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
6DHE
 
 | | RT XFEL structure of the dark-stable state of Photosystem II (0F, S1-rich) at 2.05 Angstrom resolution | | Descriptor: | (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
6DHG
 
 | | RT XFEL structure of Photosystem II 150 microseconds after the second illumination at 2.5 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
8ZN2
 
 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.65 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Bhushan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.65 A resolution.
To Be Published
|
|
8ZN3
 
 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.41 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Bhushan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.41 A resolution.
To Be Published
|
|
2ATA
 
 | | Structural Basis of DNA Recognition by p53 Tetramers (complex II) | | Descriptor: | 5'-D(*AP*AP*GP*GP*CP*AP*TP*GP*CP*CP*TP*T)-3', Cellular tumor antigen p53, ZINC ION | | Authors: | Kitayner, M, Rozenberg, H, Kessler, N, Rabinovich, D, Shakked, Z. | | Deposit date: | 2005-08-24 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of DNA Recognition by p53 Tetramers
Mol.Cell, 22, 2006
|
|
2AHI
 
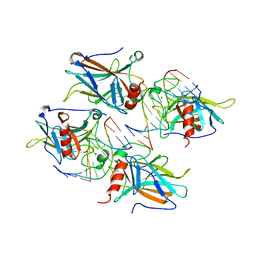 | | Structural Basis of DNA Recognition by p53 Tetramers (complex III) | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*AP*TP*GP*TP*CP*CP*G)-3', Cellular tumor antigen p53, ZINC ION | | Authors: | Kitayner, M, Rozenberg, H, Kessler, N, Rabinovich, D, Shakked, Z. | | Deposit date: | 2005-07-28 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of DNA Recognition by p53 Tetramers
Mol.Cell, 22, 2006
|
|
1EUH
 
 | | APO FORM OF A NADP DEPENDENT ALDEHYDE DEHYDROGENASE FROM STREPTOCOCCUS MUTANS | | Descriptor: | NADP DEPENDENT NON PHOSPHORYLATING GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, SULFATE ION | | Authors: | Cobessi, D, Tete-Favier, F, Marchal, S, Branlant, G, Aubry, A. | | Deposit date: | 1998-11-05 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Apo and holo crystal structures of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans.
J.Mol.Biol., 290, 1999
|
|
5VA0
 
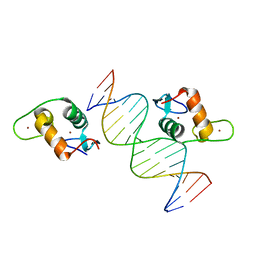 | |
3G1E
 
 | | X-ray crystal structure of coil 1A of human vimentin | | Descriptor: | Vimentin | | Authors: | Meier, M, Padilla, G.P, Herrmann, H, Wedig, T, Hergt, M, Patel, T.R, Stetefeld, J, Aebi, U, Burkhard, P. | | Deposit date: | 2009-01-29 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Vimentin coil 1A-A molecular switch involved in the initiation of filament elongation.
J.Mol.Biol., 390, 2009
|
|
5VA7
 
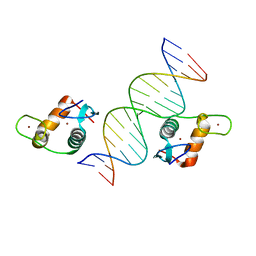 | | Glucocorticoid Receptor DNA Binding Domain - IL11 AP-1 recognition element Complex | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*TP*GP*AP*GP*TP*CP*AP*GP*GP*AP*TP*G)-3'), DNA (5'-D(*CP*AP*TP*CP*CP*TP*GP*AP*CP*TP*CP*AP*CP*CP*CP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Weikum, E.R, Ortlund, E.A. | | Deposit date: | 2017-03-24 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Tethering not required: the glucocorticoid receptor binds directly to activator protein-1 recognition motifs to repress inflammatory genes.
Nucleic Acids Res., 45, 2017
|
|
9IYF
 
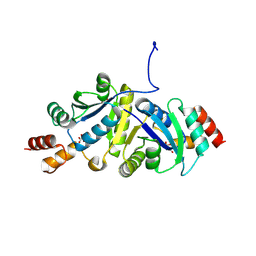 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.37 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-07-30 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.37 A resolution.
To Be Published
|
|
9IYE
 
 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.39 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-07-30 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.39 A resolution.
To Be Published
|
|
9IYH
 
 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.25 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-07-30 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.25 A resolution.
To Be Published
|
|
9IYG
 
 | |
1RAU
 
 | |
2ADY
 
 | | Structural Basis of DNA Recognition by p53 Tetramers (complex IV) | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*AP*TP*GP*TP*CP*CP*G)-3', Cellular tumor antigen p53, ZINC ION | | Authors: | Kitayner, M, Rozenberg, H, Kessler, N, Rabinovich, D, Shakked, Z. | | Deposit date: | 2005-07-21 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of DNA Recognition by p53 Tetramers
Mol.Cell, 22, 2006
|
|
2AC0
 
 | | Structural Basis of DNA Recognition by p53 Tetramers (complex I) | | Descriptor: | 5'-D(*CP*GP*GP*GP*CP*AP*TP*GP*CP*CP*CP*G)-3', Cellular tumor antigen p53, ZINC ION | | Authors: | Kitayner, M, Rozenberg, H, Kessler, N, Rabinovich, D, Shakked, Z. | | Deposit date: | 2005-07-18 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of DNA Recognition by p53 Tetramers
Mol.Cell, 22, 2006
|
|
