6CUH
 
 | | Crystal structure of the unliganded BC8B TCR | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shahine, A.E, Rossjohn, J. | | Deposit date: | 2018-03-26 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A T-cell receptor escape channel allows broad T-cell response to CD1b and membrane phospholipids.
Nat Commun, 10, 2019
|
|
3NKM
 
 | | Crystal structure of mouse autotaxin | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
3NKN
 
 | | Crystal structure of mouse autotaxin in complex with 14:0-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
5XXI
 
 | | Crystal structure of CYP2C9 in complex with multiple losartan molecules | | Descriptor: | Cytochrome P450 2C9, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Maekawa, K, Adachi, M, Shah, M.B. | | Deposit date: | 2017-07-04 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Single-Nucleotide Polymorphisms in Cytochrome P450 2C9
Biochemistry, 56, 2017
|
|
4ERT
 
 | |
4IA5
 
 | | Hydratase from Lactobacillus acidophilus - SeMet derivative (apo LAH) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Khoshnevis, S, Neumann, P, Ficner, R. | | Deposit date: | 2012-12-06 | | Release date: | 2013-03-27 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure analysis of a fatty acid double-bond hydratase from Lactobacillus acidophilus
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4IA6
 
 | | Hydratase from lactobacillus acidophilus in a ligand bound form (LA LAH) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Khoshnevis, S, Neumann, P, Ficner, R. | | Deposit date: | 2012-12-06 | | Release date: | 2013-03-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure analysis of a fatty acid double-bond hydratase from Lactobacillus acidophilus
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7PO4
 
 | | Assembly intermediate of human mitochondrial ribosome large subunit (largely unfolded rRNA with MALSU1, L0R8F8 and ACP) | | Descriptor: | 16SrRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Itoh, Y, Khawaja, A, Rorbach, J, Amunts, A. | | Deposit date: | 2021-09-08 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Mechanism of mitoribosomal small subunit biogenesis and preinitiation.
Nature, 606, 2022
|
|
7PNY
 
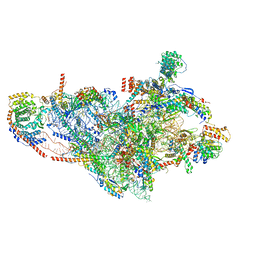 | | Assembly intermediate of human mitochondrial ribosome small subunit without mS37 in complex with RBFA and METTL15 conformation b | | Descriptor: | 12S mitochondrial rRNA, 12S rRNA N4-methylcytidine (m4C) methyltransferase, 28S ribosomal protein S10, ... | | Authors: | Itoh, Y, Khawaja, A, Rorbach, J, Amunts, A. | | Deposit date: | 2021-09-08 | | Release date: | 2022-06-15 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Mechanism of mitoribosomal small subunit biogenesis and preinitiation.
Nature, 606, 2022
|
|
7PO3
 
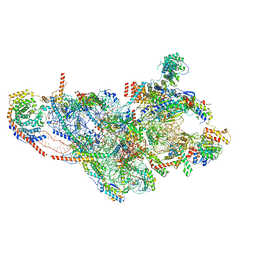 | | Human mitochondrial ribosome small subunit | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Itoh, Y, Khawaja, A, Rorbach, J, Amunts, A. | | Deposit date: | 2021-09-08 | | Release date: | 2022-06-15 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Mechanism of mitoribosomal small subunit biogenesis and preinitiation.
Nature, 606, 2022
|
|
7PNZ
 
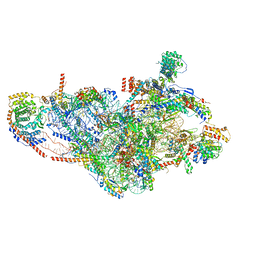 | | Assembly intermediate of human mitochondrial ribosome small subunit without mS37 in complex with RBFA and METTL15 conformation c | | Descriptor: | 12S mitochondrial rRNA, 12S rRNA N4-methylcytidine (m4C) methyltransferase, 28S ribosomal protein S10, ... | | Authors: | Itoh, Y, Khawaja, A, Rorbach, J, Amunts, A. | | Deposit date: | 2021-09-08 | | Release date: | 2022-06-15 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Mechanism of mitoribosomal small subunit biogenesis and preinitiation.
Nature, 606, 2022
|
|
7PO0
 
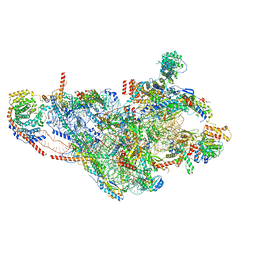 | | Assembly intermediate of human mitochondrial ribosome small subunit without mS37 in complex with RBFA and IF3 | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Itoh, Y, Khawaja, A, Rorbach, J, Amunts, A. | | Deposit date: | 2021-09-08 | | Release date: | 2022-06-15 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of mitoribosomal small subunit biogenesis and preinitiation.
Nature, 606, 2022
|
|
7PNT
 
 | | Assembly intermediate of mouse mitochondrial ribosome small subunit without mS37 in complex with RbfA and Tfb1m | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Itoh, Y, Khawaja, A, Laptev, I, Sergiev, P, Rorbach, J, Amunts, A. | | Deposit date: | 2021-09-08 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Mechanism of mitoribosomal small subunit biogenesis and preinitiation.
Nature, 606, 2022
|
|
4EK1
 
 | | Crystal Structure of Electron-Spin Labeled Cytochrome P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Goodin, D.B. | | Deposit date: | 2012-04-08 | | Release date: | 2012-07-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Double electron-electron resonance shows cytochrome P450cam undergoes a conformational change in solution upon binding substrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5X23
 
 | | Crystal structure of CYP2C9 genetic variant A477T (*30) in complex with multiple losartan molecules | | Descriptor: | Cytochrome P450 2C9, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Maekawa, K, Adachi, M, Shah, M.B. | | Deposit date: | 2017-01-30 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Single-Nucleotide Polymorphisms in Cytochrome P450 2C9
Biochemistry, 56, 2017
|
|
5X24
 
 | | Crystal structure of CYP2C9 genetic variant I359L (*3) in complex with multiple losartan molecules | | Descriptor: | Cytochrome P450 2C9, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Maekawa, K, Adachi, M, Shah, M.B. | | Deposit date: | 2017-01-30 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural Basis of Single-Nucleotide Polymorphisms in Cytochrome P450 2C9
Biochemistry, 56, 2017
|
|
3BXL
 
 | | Crystal structure of the R-type calcium channeL (CaV2.3) IQ domain and CA2+calmodulin complex | | Descriptor: | CALCIUM ION, Calmodulin, SULFATE ION, ... | | Authors: | Mori, M.X, Vander Kooi, C.W, Leahy, D.J, Yue, D.T. | | Deposit date: | 2008-01-14 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the CaV2 IQ domain in complex with Ca2+/calmodulin
To be Published
|
|
2IZQ
 
 | | Gramicidin D complex with KI | | Descriptor: | GRAMICIDIN D, IODIDE ION, METHANOL, ... | | Authors: | Olczak, A, Glowka, M.L, Szczesio, M, Bojarska, J, Duax, W.L, Burkhart, B.M, Wawrzak, Z. | | Deposit date: | 2006-07-26 | | Release date: | 2007-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Nonstoichiometric Complex of Gramicidin D with Ki at 0.80 A Resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4JB1
 
 | | Crystal structure of P. aeruginosa MurB in complex with NADP+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Chen, M.W, Lohkamp, B, Schnell, R, Lescar, J, Schneider, G. | | Deposit date: | 2013-02-19 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate Channel Flexibility in Pseudomonas aeruginosa MurB Accommodates Two Distinct Substrates.
Plos One, 8, 2013
|
|
3DVE
 
 | | Crystal Structure of Ca2+/CaM-CaV2.2 IQ domain complex | | Descriptor: | CALCIUM ION, Calmodulin, NICKEL (II) ION, ... | | Authors: | Kim, E.Y, Rumpf, C.H, Fujiwara, Y, Cooley, E.S, Van Petegem, F, Minor, D.L. | | Deposit date: | 2008-07-18 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of Ca(V)2 Ca(2+)/CaM-IQ Domain Complexes Reveal Binding Modes that Underlie Calcium-Dependent Inactivation and Facilitation.
Structure, 16, 2008
|
|
3DVM
 
 | | Crystal Structure of Ca2+/CaM-CaV2.1 IQ domain complex | | Descriptor: | CALCIUM ION, Calmodulin, Voltage-dependent P/Q-type calcium channel subunit alpha-1A | | Authors: | Kim, E.Y, Rumpf, C.H, Fujiwara, Y, Cooley, E.S, Van Petegem, F, Minor, D.L. | | Deposit date: | 2008-07-18 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of Ca(V)2 Ca(2+)/CaM-IQ Domain Complexes Reveal Binding Modes that Underlie Calcium-Dependent Inactivation and Facilitation.
Structure, 16, 2008
|
|
3DVK
 
 | | Crystal Structure of Ca2+/CaM-CaV2.3 IQ domain complex | | Descriptor: | CALCIUM ION, Calmodulin, Voltage-dependent R-type calcium channel subunit alpha-1E | | Authors: | Kim, E.Y, Rumpf, C.H, Fujiwara, Y, Cooley, E.S, Van Petegem, F, Minor, D.L. | | Deposit date: | 2008-07-18 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Ca(V)2 Ca(2+)/CaM-IQ Domain Complexes Reveal Binding Modes that Underlie Calcium-Dependent Inactivation and Facilitation.
Structure, 16, 2008
|
|
3DVJ
 
 | | Crystal Structure of Ca2+/CaM-CaV2.2 IQ domain (without cloning artifact, HM to TV) complex | | Descriptor: | CALCIUM ION, Calmodulin, Voltage-dependent N-type calcium channel subunit alpha-1B | | Authors: | Kim, E.Y, Rumpf, C.H, Fujiwara, Y, Cooley, E.S, Van Petegem, F, Minor, D.L. | | Deposit date: | 2008-07-18 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Ca(V)2 Ca(2+)/CaM-IQ Domain Complexes Reveal Binding Modes that Underlie Calcium-Dependent Inactivation and Facilitation.
Structure, 16, 2008
|
|
2LZO
 
 | | Spatial structure of Pi-AnmTX Ugr 9a-1 | | Descriptor: | UGTX | | Authors: | Mineev, K, Arseniev, A. | | Deposit date: | 2012-10-08 | | Release date: | 2013-07-03 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Sea Anemone Peptide with Uncommon beta-Hairpin Structure Inhibits Acid-sensing Ion Channel 3 (ASIC3) and Reveals Analgesic Activity.
J.Biol.Chem., 288, 2013
|
|
1XTA
 
 | | Crystal Structure of Natrin, a snake venom CRISP from Taiwan cobra (Naja atra) | | Descriptor: | Natrin 1 | | Authors: | Wang, Y.-L, Goh, K.-X, Lee, S.-C, Huang, W.-N, Wu, W.-G, Chen, C.-J. | | Deposit date: | 2004-10-21 | | Release date: | 2005-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structures of snake venom CRISP reveal an action mechanism involving serine protease and ion channel blocking domains
To be Published
|
|
