7KIN
 
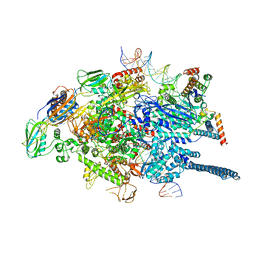 | | Mycobacterium tuberculosis WT RNAP transcription open promoter complex with WhiB7 promoter | | Descriptor: | DNA (49-MER), DNA (54-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-10-23 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural basis of transcriptional activation by the Mycobacterium tuberculosis intrinsic antibiotic-resistance transcription factor WhiB7.
Mol.Cell, 81, 2021
|
|
7KIF
 
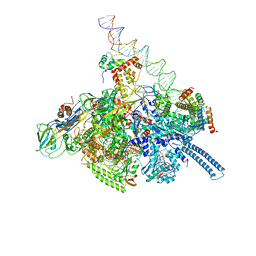 | | Mycobacterium tuberculosis WT RNAP transcription open promoter complex with WhiB7 transcription factor | | Descriptor: | DNA (55-MER), DNA (63-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-10-23 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis of transcriptional activation by the Mycobacterium tuberculosis intrinsic antibiotic-resistance transcription factor WhiB7.
Mol.Cell, 81, 2021
|
|
7KIM
 
 | | Mycobacterium tuberculosis WT RNAP transcription closed promoter complex with WhiB7 transcription factor | | Descriptor: | DNA (45-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-10-23 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis of transcriptional activation by the Mycobacterium tuberculosis intrinsic antibiotic-resistance transcription factor WhiB7.
Mol.Cell, 81, 2021
|
|
5LDI
 
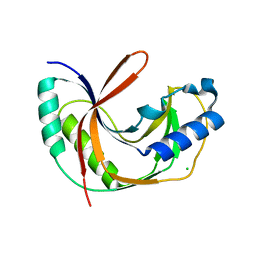 | | Crystal structure of E.coli LigT in apo form | | Descriptor: | CHLORIDE ION, RNA 2',3'-cyclic phosphodiesterase | | Authors: | Myllykoski, M, Kursula, P. | | Deposit date: | 2016-06-27 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural aspects of nucleotide ligand binding by a bacterial 2H phosphoesterase.
PLoS ONE, 12, 2017
|
|
5LDJ
 
 | |
6L3F
 
 | |
8APX
 
 | |
7ORI
 
 | | La Crosse virus polymerase at replication late-elongation stage | | Descriptor: | La Crosse virus polymerase, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
8E74
 
 | | Mycobacterium tuberculosis RNAP paused elongation complex with NusG transcription factor | | Descriptor: | DNA (54-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Delbeau, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2022-08-23 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural and functional basis of the universal transcription factor NusG pro-pausing activity in Mycobacterium tuberculosis.
Mol.Cell, 83, 2023
|
|
8E8M
 
 | | Mycobacterium tuberculosis RNAP paused elongation complex | | Descriptor: | DNA (54-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Delbeau, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural and functional basis of the universal transcription factor NusG pro-pausing activity in Mycobacterium tuberculosis.
Mol.Cell, 83, 2023
|
|
8E82
 
 | | Mycobacterium tuberculosis RNAP elongation complex with NusG transcription factor | | Descriptor: | DNA (54-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Delbeau, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural and functional basis of the universal transcription factor NusG pro-pausing activity in Mycobacterium tuberculosis.
Mol.Cell, 83, 2023
|
|
8E79
 
 | | Mycobacterium tuberculosis RNAP paused elongation complex with Escherichia coli NusG transcription factor | | Descriptor: | DNA (54-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Delbeau, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2022-08-23 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural and functional basis of the universal transcription factor NusG pro-pausing activity in Mycobacterium tuberculosis.
Mol.Cell, 83, 2023
|
|
8E95
 
 | | Mycobacterium tuberculosis RNAP elongation complex | | Descriptor: | DNA (25-MER), DNA (33-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Delbeau, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and functional basis of the universal transcription factor NusG pro-pausing activity in Mycobacterium tuberculosis.
Mol.Cell, 83, 2023
|
|
4X9C
 
 | | 1.4A crystal structure of Hfq from Methanococcus jannaschii | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nikulin, A.D, Tishchenko, S.V, Nikonova, S.V, Murina, V.N, Mihailina, A.O, Lekontseva, N.V. | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterization of RNA-binding properties of the archaeal Hfq-like protein from Methanococcus jannaschii.
J. Biomol. Struct. Dyn., 35, 2017
|
|
6FKR
 
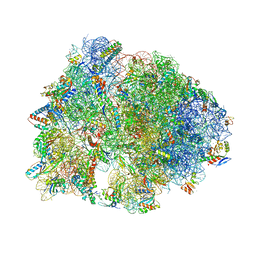 | | Crystal structure of the dolphin proline-rich antimicrobial peptide Tur1A bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16 ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mardirossian, M, Perebaskine, N, Benincasa, M, Gambato, S, Hofmann, S, Huter, P, Muller, C, Hilpert, K, Innis, C.A, Tossi, A, Wilson, D.N. | | Deposit date: | 2018-01-24 | | Release date: | 2018-03-28 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Dolphin Proline-Rich Antimicrobial Peptide Tur1A Inhibits Protein Synthesis by Targeting the Bacterial Ribosome.
Cell Chem Biol, 25, 2018
|
|
5LDQ
 
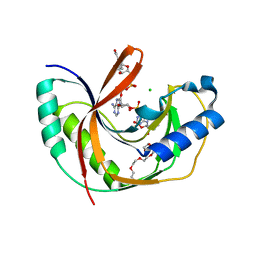 | | Crystal structure of E.coli LigT complexed with NADP+ | | Descriptor: | CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PENTAETHYLENE GLYCOL, ... | | Authors: | Myllykoski, M, Kursula, P. | | Deposit date: | 2016-06-27 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural aspects of nucleotide ligand binding by a bacterial 2H phosphoesterase.
PLoS ONE, 12, 2017
|
|
5LDM
 
 | |
5LDK
 
 | |
5LDP
 
 | |
6WAZ
 
 | | +1 extended HIV-1 reverse transcriptase initiation complex core (pre-translocation state) | | Descriptor: | HIV-1 viral RNA genome fragment, Reverse transcriptase p51 subunit, Reverse transcriptase/ribonuclease H, ... | | Authors: | Larsen, K.P, Jackson, L.N, Kappel, K, Zhang, J, Puglisi, E.V. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.
J.Mol.Biol., 432, 2020
|
|
6D1R
 
 | | Structure of Staphylococcus aureus RNase P protein at 2.0 angstrom | | Descriptor: | Ribonuclease P protein component | | Authors: | Ha, L, Colquhoun, J, Noinaj, N, Das, C, Dunman, P, Flaherty, D.P. | | Deposit date: | 2018-04-12 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Crystal structure of the ribonuclease-P-protein subunit from Staphylococcus aureus.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5BTE
 
 | |
1T4O
 
 | | Crystal structure of rnt1p dsRBD | | Descriptor: | Ribonuclease III | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Graille, M, van Tilbeurgh, H, Leeper, T.C, Godin, K.S, Edwards, T.E, Sigurdsson, S.T, Rozenkrants, N, Nagel, R.J, Ares Jr, M, Varani, G. | | Deposit date: | 2004-04-30 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new alpha-helical extension promotes RNA binding by the dsRBD of Rnt1p RNAse III
Embo J., 23, 2004
|
|
1T4N
 
 | | Solution structure of Rnt1p dsRBD | | Descriptor: | Ribonuclease III | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Graille, M, van Tilbeurgh, H, Leeper, T.C, Godin, K.S, Edwards, T.E, Sigurdsson, S.T, Rozenkrants, N, Nagel, R.J, Ares, M, Varani, G. | | Deposit date: | 2004-04-30 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A new alpha-helical extension promotes RNA binding by the dsRBD of Rnt1p RNAse III
Embo J., 23, 2004
|
|
6WB2
 
 | | +3 extended HIV-1 reverse transcriptase initiation complex core (displaced state) | | Descriptor: | HIV-1 viral RNA genome fragment, Reverse transcriptase/ribonuclease H, reverse transcriptase p51 subunit, ... | | Authors: | Larsen, K.P, Jackson, L.N, Kappel, K, Zhang, J, Chen, D.H, Puglisi, E.V. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.
J.Mol.Biol., 432, 2020
|
|
