8Z9Q
 
 | | Cryo-EM structure of Thogoto virus polymerase in a replication reception conformation | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-D(*(ATP))-R(P*GP*CP*AP*AP*AP*AP*AP*CP*A)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8BYQ
 
 | | RNA polymerase II pre-initiation complex with the proximal +1 nucleosome (PIC-Nuc10W) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
8BVW
 
 | | RNA polymerase II pre-initiation complex with the distal +1 nucleosome (PIC-Nuc18W) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-20 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
8G5Z
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G6J
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state 2) | | Descriptor: | (3R,6R,9S,12S,15S,18S,20R,24aR)-6-[(2S)-butan-2-yl]-3,12-bis[(1R)-1-hydroxy-2-methylpropyl]-8,9,11,17,18-pentamethyl-15-[(2S)-2-methylbutyl]hexadecahydropyrido[1,2-a][1,4,7,10,13,16,19]heptaazacyclohenicosine-1,4,7,10,13,16,19(21H)-heptone, (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, 1,4-DIAMINOBUTANE, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-15 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G60
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (CR state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G61
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (AC state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8GLP
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (Consensus LSU focused refined structure) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Watson, Z.L, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-03-22 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (1.67 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G5Y
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (IC state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
7NKX
 
 | | RNA polymerase II-Spt4/5-nucleosome-Chd1 structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromatin elongation factor SPT4, ... | | Authors: | Farnung, L, Ochmann, M, Engeholm, M, Cramer, P. | | Deposit date: | 2021-02-19 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome transcription mediated by Chd1 and FACT.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8BZ1
 
 | | RNA polymerase II core pre-initiation complex with the proximal +1 nucleosome (cPIC-Nuc10W) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
7Z1L
 
 | | Structure of yeast RNA Polymerase III Pre-Termination Complex (PTC) | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1O
 
 | | Structure of yeast RNA Polymerase III PTC + NTPs | | Descriptor: | CHAPSO, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1M
 
 | | Structure of yeast RNA Polymerase III Elongation Complex (EC) | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1N
 
 | | Structure of yeast RNA Polymerase III Delta C53-C37-C11 | | Descriptor: | CHAPSO, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
2E2J
 
 | | RNA polymerase II elongation complex in 5 mM Mg+2 with GMPCPP | | Descriptor: | 27-MER DNA template strand, 5'-D(P*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*A)-3', 5'-R(P*AP*UP*CP*GP*AP*GP*AP*GP*G)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
2E2H
 
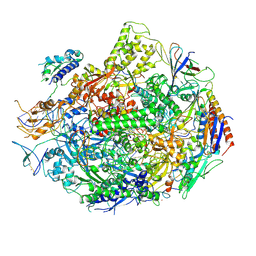 | | RNA polymerase II elongation complex at 5 mM Mg2+ with GTP | | Descriptor: | 28-MER DNA template strand, 5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3', 5'-R(*AP*UP*CP*GP*AP*GP*AP*GP*GP*A)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
7F4G
 
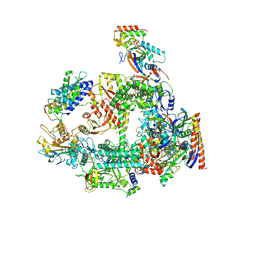 | | Structure of RPAP2-bound RNA polymerase II | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Chen, X, Qi, Y, Wang, X, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-06-18 | | Release date: | 2021-07-07 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | RPAP2 regulates a transcription initiation checkpoint by inhibiting assembly of pre-initiation complex.
Cell Rep, 39, 2022
|
|
2E2I
 
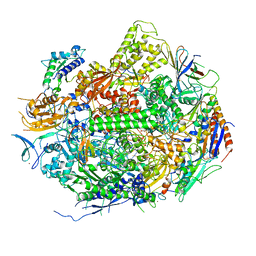 | | RNA polymerase II elongation complex in 5 mM Mg+2 with 2'-dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 28-MER DNA template strand, 5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
6T0R
 
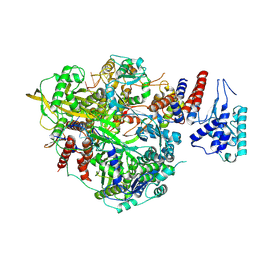 | | Bat Influenza A polymerase product dissociation complex using 44-mer vRNA template with mutated oligo(U) sequence | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA-directed RNA polymerase catalytic subunit, ... | | Authors: | Wandzik, J.M, Kouba, T, Cusack, S. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase.
Cell, 181, 2020
|
|
3S2D
 
 | | RNA Polymerase II Initiation Complex with a 5-nt RNA containing a 5Br-U | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*AP*CP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3S1Q
 
 | | RNA Polymerase II Initiation Complex with a 5-nt 3'-deoxy RNA soaked with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3S15
 
 | | RNA Polymerase II Initiation Complex with a 7-nt RNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-14 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3S2H
 
 | | RNA Polymerase II Initiation Complex with a 6-nt RNA containing a 2[prime]-iodo ATP | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3S1M
 
 | | RNA Polymerase II Initiation Complex with a 5-nt RNA (variant 1) | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*GP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-15 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
