7SWD
 
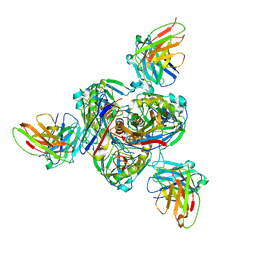 | | Structure of EBOV GP lacking the mucin-like domain with 1C11 scFv and 1C3 Fab bound | | Descriptor: | 1C11 scFv, 1C3 heavy chain, 1C3 light chain, ... | | Authors: | Milligan, J.C, Yu, X, Saphire, E.O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-04-06 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Asymmetric and non-stoichiometric glycoprotein recognition by two distinct antibodies results in broad protection against ebolaviruses.
Cell, 185, 2022
|
|
7ZX9
 
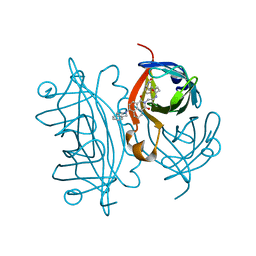 | | Streptavidin with a fluorescent substrate | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[2-[6-(3,4-dihydro-1~{H}-isoquinolin-2-yl)-1,3-bis(oxidanylidene)benzo[de]isoquinolin-2-yl]ethyl]pentanamide, Streptavidin | | Authors: | Igareta, N.V, Ward, T.R. | | Deposit date: | 2022-05-20 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.546 Å) | | Cite: | Streptavidin Iron-Porphyrin
To Be Published
|
|
7ZOF
 
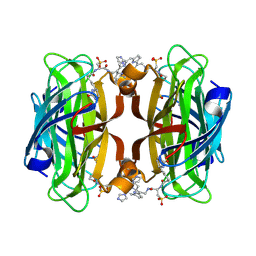 | | Streptavidin Iron-Porphyrin | | Descriptor: | (2R)-2-[5-[(3aS,4S,6aR)-2-oxidanylidene-1,3,3a,4,6,6a-hexahydrothieno[3,4-d]imidazol-4-yl]pentanoylamino]-3-[[(5Z,10Z,14Z,19Z)-15-[[[(2R)-2-[5-[(3aS,4S,6aR)-2-oxidanylidene-1,3,3a,4,6,6a-hexahydrothieno[3,4-d]imidazol-4-yl]pentanoylamino]-3-sulfo-propanoyl]amino]methyl]-1,4,21,23-tetrahydroporphyrin-5-yl]methylamino]-3-oxidanylidene-propane-1-sulfonic acid, FE (III) ION, IMIDAZOLE, ... | | Authors: | Igareta, N.V, Ward, T.R. | | Deposit date: | 2022-04-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Streptavidin Iron-Porphyrin
To Be Published
|
|
4YC1
 
 | |
6QJJ
 
 | |
5IHN
 
 | |
6QJD
 
 | |
6QJN
 
 | |
5IHI
 
 | |
5I11
 
 | |
7D6Q
 
 | | Crystal structure of the Stx2a | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, Shiga toxin 2 B subunit, rRNA N-glycosylase | | Authors: | Takahashi, M, Tamada, M, Hibino, M, Senda, M, Okuda, A, Miyazawa, A, Senda, T, Nishikawa, K. | | Deposit date: | 2020-10-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a peptide motif that potently inhibits two functionally distinct subunits of Shiga toxin.
Commun Biol, 4, 2021
|
|
7D6R
 
 | | Crystal structure of the Stx2a complexed with MMA betaAla peptide | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, MMA betaAla peptide, Shiga toxin 2 B subunit, ... | | Authors: | Takahashi, M, Tamada, M, Hibino, M, Senda, M, Okuda, A, Miyazawa, A, Senda, T, Nishikawa, K. | | Deposit date: | 2020-10-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a peptide motif that potently inhibits two functionally distinct subunits of Shiga toxin.
Commun Biol, 4, 2021
|
|
6QJI
 
 | |
4Y92
 
 | |
5IHK
 
 | |
6QJF
 
 | |
6QJL
 
 | |
6QJG
 
 | |
6QJK
 
 | |
3NAN
 
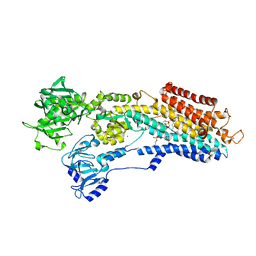 | | SR Ca(2+)-ATPase in the HnE2 state complexed with a Thapsigargin derivative Boc-(phi)Tg | | Descriptor: | (3S,3aR,4S,6S,6aR,7S,8S,9R,9aS,9bS)-6-(acetyloxy)-4-{[4-(3-{6-[(tert-butoxycarbonyl)amino]hexyl}-4-hydroxyphenyl)butanoyl]oxy}-3,3a-dihydroxy-3,6,9-trimethyl-8-{[(2Z)-2-methylbut-2-enoyl]oxy}-2-oxododecahydroazuleno[4,5-b]furan-7-yl octanoate, MAGNESIUM ION, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Winther, A.M.L, Sonntag, Y, Olesen, C, Moller, J.V, Nissen, P. | | Deposit date: | 2010-06-02 | | Release date: | 2010-06-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Critical roles of hydrophobicity and orientation of side chains for inactivation of sarcoplasmic reticulum Ca2+-ATPase with thapsigargin and thapsigargin analogs
J.Biol.Chem., 285, 2010
|
|
3NAL
 
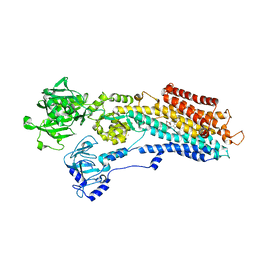 | | SR Ca(2+)-ATPase in the HnE2 state complexed with the Thapsigargin derivative DTB | | Descriptor: | (3S,3aR,4S,6S,6aS,8R,9bS)-6-(acetyloxy)-3,3a-dihydroxy-3,6,9-trimethyl-8-{[(2Z)-2-methylbut-2-enoyl]oxy}-2-oxo-2,3,3a,4,5,6,6a,7,8,9b-decahydroazuleno[4,5-b]furan-4-yl dodecanoate, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Winther, A.M.L, Sonntag, Y, Olesen, C, Moller, J.V, Nissen, P. | | Deposit date: | 2010-06-02 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Critical roles of hydrophobicity and orientation of side chains for inactivation of sarcoplasmic reticulum Ca2+-ATPase with thapsigargin and thapsigargin analogs
J.Biol.Chem., 285, 2010
|
|
3NAM
 
 | | SR Ca(2+)-ATPase in the HnE2 state complexed with the Thapsigargin derivative dOTg | | Descriptor: | (3S,3aR,4S,6S,6aS,8R,9R,9aR,9bS)-6-(acetyloxy)-4-(butanoyloxy)-3,3a-dihydroxy-3,6,9-trimethyl-2-oxododecahydroazuleno[4,5-b]furan-8-yl (2Z)-2-methylbut-2-enoate, MAGNESIUM ION, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Winther, A.M.L, Sonntag, Y, Olesen, C, Moller, J.V, Nissen, P. | | Deposit date: | 2010-06-02 | | Release date: | 2010-06-30 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Critical roles of hydrophobicity and orientation of side chains for inactivation of sarcoplasmic reticulum Ca2+-ATPase with thapsigargin and thapsigargin analogs
J.Biol.Chem., 285, 2010
|
|
6IEI
 
 | |
6IDC
 
 | |
6IYS
 
 | |
