5TKG
 
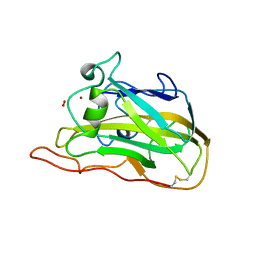 | | Neurospora crassa polysaccharide monooxygenase 2 resting state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Lytic polysaccharide monooxygenase, ... | | Authors: | O'Dell, W.B, Meilleur, F. | | Deposit date: | 2016-10-06 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Oxygen Activation at the Active Site of a Fungal Lytic Polysaccharide Monooxygenase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8Q3S
 
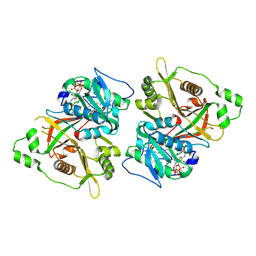 | |
5TH7
 
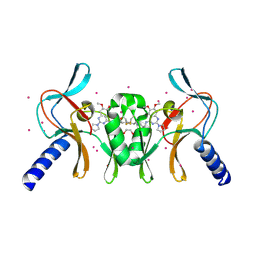 | | Complex of SETD8 with MS453 | | Descriptor: | 1,2-ETHANEDIOL, N-(3-{[6,7-dimethoxy-2-(pyrrolidin-1-yl)quinazolin-4-yl]amino}propyl)propanamide, N-lysine methyltransferase KMT5A, ... | | Authors: | Yu, W, Tempel, W, Babault, N, Ma, A, Butler, K.V, Jin, J, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Design of a Covalent Inhibitor of the SET Domain-Containing Protein 8 (SETD8) Lysine Methyltransferase.
J. Med. Chem., 59, 2016
|
|
7UXP
 
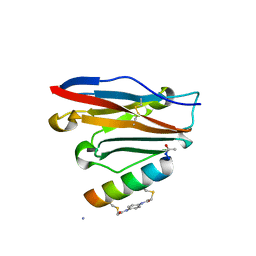 | | Structure of PDL1 in complex with FP28132, a Helicon Polypeptide | | Descriptor: | AMINO GROUP, FP28132, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Travaline, T, Tattersfield, H, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6MEF
 
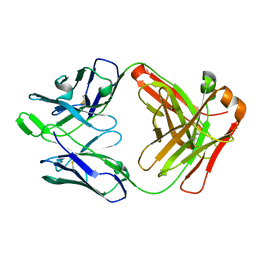 | | Crystal structure of broadly neutralizing antibody AR3C | | Descriptor: | antibody AR3C Heavy Chain, antibody AR3C Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J. | | Deposit date: | 2018-09-06 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | HCV Broadly Neutralizing Antibodies Use a CDRH3 Disulfide Motif to Recognize an E2 Glycoprotein Site that Can Be Targeted for Vaccine Design.
Cell Host Microbe, 24, 2018
|
|
5THA
 
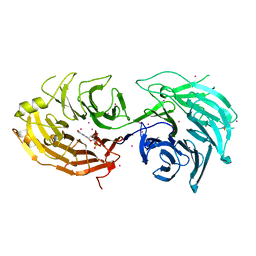 | | Gemin5 WD40 repeats in complex with a guanosyl moiety | | Descriptor: | DIGUANOSINE-5'-TRIPHOSPHATE, Gem-associated protein 5, UNKNOWN ATOM OR ION | | Authors: | Chao, X, Tempel, W, Bian, C, Cerovina, T, He, H, Seitova, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-29 | | Release date: | 2016-10-19 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly.
Genes Dev., 30, 2016
|
|
7OJD
 
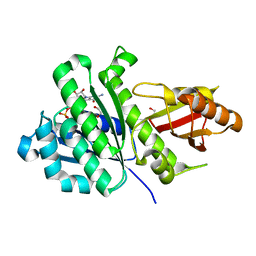 | | SaFtsZ(D46A) complexed with GDP | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-14 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
8TTD
 
 | |
7OMP
 
 | | SaFtsZ complexed with GDPPCP and Mg2+ | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, MAGNESIUM ION, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7UXI
 
 | | Structure of CDK2 in complex with FP19711, a Helicon Polypeptide | | Descriptor: | AMINO GROUP, Cyclin-dependent kinase 2, FP19711, ... | | Authors: | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6MES
 
 | |
7UX5
 
 | | Structure of PDL1 in complex with FP28136, a Helicon Polypeptide | | Descriptor: | Helicon FP28136, N,N'-(1,4-phenylene)diacetamide, Programmed cell death 1 ligand 1 | | Authors: | Agarwal, S, Li, K, Tokareva, O, Thomson, T, Travaline, T, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7MQY
 
 | | Zebrafish CNTN4 APLP2 complex | | Descriptor: | Fusion protein of CNTN4 and APLP2 | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-07 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
5THT
 
 | | Crystal Structure of G303A HDAC8 in complex with M344 | | Descriptor: | 1,2-ETHANEDIOL, 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Histone deacetylase 8, ... | | Authors: | Porter, N.J, Christianson, D.W. | | Deposit date: | 2016-09-30 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Structural and Functional Influence of the Glycine-Rich Loop G302GGGY on the Catalytic Tyrosine of Histone Deacetylase 8.
Biochemistry, 55, 2016
|
|
7OJA
 
 | | SaFtsZ(D210N) complexed with GDP | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-14 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OM5
 
 | | Anti-EGFR nanobody EgB4 | | Descriptor: | GLYCEROL, Nanobody EgB4, ZINC ION | | Authors: | Zeronian, M.R, Janssen, B.J.C. | | Deposit date: | 2021-05-21 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural insights into the non-inhibitory mechanism of the anti-EGFR EgB4 nanobody.
Bmc Mol Cell Biol, 23, 2022
|
|
6MEI
 
 | |
8JMS
 
 | |
7UXM
 
 | | Structure of PPIA in complex with FP29092, a Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, AMINO GROUP, FP29092, ... | | Authors: | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7MRO
 
 | | Zebrafish CNTN4 FN1-FN3 domains | | Descriptor: | Contactin-4 | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-07 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
7ON4
 
 | | SaFtsZ complexed with GDP (co-crystalization with 1mM EDTA) | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
8TYG
 
 | | Plasmodium vivax PMV-WM08 inhibitor complex | | Descriptor: | (2E,4aR,7aS)-6-[3-(4-chlorophenyl)pyridin-2-yl]-7a-(2,5-difluorophenyl)-2-imino-3-methyloctahydro-4H-pyrrolo[3,4-d]pyrimidin-4-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hodder, A.N, Scally, S.W, Cowman, A.F. | | Deposit date: | 2023-08-25 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Plasmodium vivax PMV-WM08 inhibitor complex
To Be Published
|
|
7MRS
 
 | | Zebrafish CNTN4 APPb complex | | Descriptor: | Amyloid-beta A4 protein, Contactin-4 | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-09 | | Release date: | 2022-01-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
8TYH
 
 | | Plasmodium vivax PMX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-tungstotellurate(VI), Aspartyl protease, ... | | Authors: | Hodder, A.N, Scally, S.W, Cowman, A.F. | | Deposit date: | 2023-08-25 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Plasmodium vivax PMX
To Be Published
|
|
6MEN
 
 | | Crystal structure of a Tylonycteris bat coronavirus HKU4 macrodomain in complex with adenosine diphosphate glucose (ADP-glucose) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE-GLUCOSE, Replicase polyprotein 1ab | | Authors: | Hammond, R.G, Schormann, N, McPherson, R.L, Leung, A.K.L, Deivanayagam, C.C.S, Johnson, M.A. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | ADP-Ribose and Analogues bound to the DeMARylating Macrodomain from the Bat Coronavirus HKU4
Proc.Natl.Acad.Sci.USA, 2021
|
|
