5EP0
 
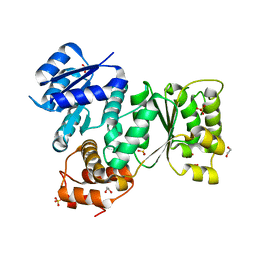 | | Quorum-Sensing Signal Integrator LuxO - Receiver+Catalytic Domains | | Descriptor: | 1,2-ETHANEDIOL, Putative repressor protein luxO, SULFATE ION | | Authors: | Shah, T, Selcuk, H.B, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2015-11-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO.
Plos Biol., 14, 2016
|
|
7QBA
 
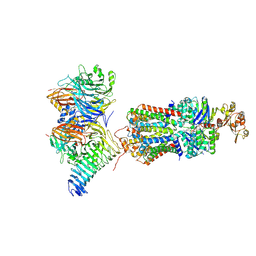 | | CryoEM structure of the ABC transporter NosDFY complexed with nitrous oxide reductase NosZ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Zipfel, S, Mueller, C, Topitsch, A, Lutz, M, Zhang, L, Einsle, O. | | Deposit date: | 2021-11-18 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7Q1N
 
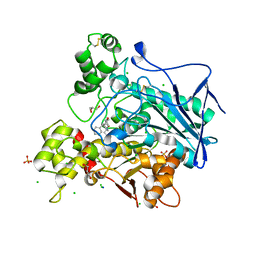 | | Crystal structure of human butyrylcholinesterase in complex with N-[(2R)-3-[(cyclohexylmethyl)amino]-2-hydroxypropyl]-2,2-diphenylacetamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Brazzolotto, X, Panek, D, Pasieka, A, Malawska, B, Nachon, F. | | Deposit date: | 2021-10-20 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of new, highly potent and selective inhibitors of BuChE - design, synthesis, in vitro and in vivo evaluation and crystallography studies.
Eur.J.Med.Chem., 249, 2023
|
|
7Q6M
 
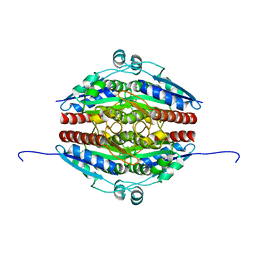 | | Structure of WrbA from Yersinia pseudotuberculosis in P1 | | Descriptor: | CHLORIDE ION, NAD(P)H dehydrogenase (quinone) | | Authors: | Gabrielsen, M, Beckham, K.S.H, Roe, A.J. | | Deposit date: | 2021-11-08 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structures of WrbA, a spurious target of the salicylidene acylhydrazide inhibitors of type III secretion in Gram-negative pathogens, and verification of improved specificity of next-generation compounds.
Microbiology (Reading, Engl.), 168, 2022
|
|
5EIF
 
 | |
3MEG
 
 | | HIV-1 K103N Reverse Transcriptase in Complex with TMC278 | | Descriptor: | 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, SULFATE ION, p51 Reverse transcriptase, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase with Etravirine (TMC125) and Rilpivirine (TMC278): Implications for Drug Design.
J.Med.Chem., 53, 2010
|
|
6OOP
 
 | | protein B | | Descriptor: | 1,1'-dimethyl-4,4'-bipyridin-1-ium, Multidrug transporter MdfA, PRASEODYMIUM ION | | Authors: | Lu, M. | | Deposit date: | 2019-04-23 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an engineered multidrug transporter MdfA reveals the molecular basis for substrate recognition.
Commun Biol, 2, 2019
|
|
5EQG
 
 | | Human GLUT1 in complex with inhibitor (2~{S})-3-(4-fluorophenyl)-2-[2-(3-hydroxyphenyl)ethanoylamino]-~{N}-[(1~{S})-1-phenylethyl]propanamide | | Descriptor: | (2~{S})-3-(4-fluorophenyl)-2-[2-(3-hydroxyphenyl)ethanoylamino]-~{N}-[(1~{S})-1-phenylethyl]propanamide, Solute carrier family 2, facilitated glucose transporter member 1 | | Authors: | Kapoor, K, Finer-Moore, J, Pedersen, B.P, Caboni, L, Waight, A.B, Hillig, R, Bringmann, P, Heisler, I, Muller, T, Siebeneicher, H, Stroud, R.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of inhibition of human glucose transporter GLUT1 is conserved between cytochalasin B and phenylalanine amides.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4AQF
 
 | | X-ray crystallographic structure of Crimean-congo haemorrhagic fever virus nucleoprotein | | Descriptor: | NUCLEOPROTEIN, SULFATE ION | | Authors: | Wang, Y, Dutta, S, Karlberg, H, Devignot, S, Weber, F, Hao, Q, Tan, Y.J, Mirazimi, A, Kotaka, M. | | Deposit date: | 2012-04-17 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Crimean-Congo Haemorraghic Fever Virus Nucleoprotein: Superhelical Homo-Oligomers and the Role of Caspase-3 Cleavage.
J.Virol., 86, 2012
|
|
7ALX
 
 | | Sav-SOD: Chimeric Streptavidin-cSOD as Host for Artificial Metalloenzymes | | Descriptor: | ((4S)-1,3-dimesityl-4-((5-((3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)pentanamido)methyl)imidazolidin-2-yl)gold, Streptavidin,Superoxide dismutase [Cu-Zn],Streptavidin | | Authors: | Igareta, N.V, Christoffel, F, Peterson, R.L, Ward, T.R. | | Deposit date: | 2020-10-07 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and evolution of chimeric streptavidin for protein-enabled dual gold catalysis
Nat Catal, 4, 2021
|
|
6OOM
 
 | | PROTEIN A | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, Multidrug transporter MdfA, PRASEODYMIUM ION | | Authors: | Lu, M. | | Deposit date: | 2019-04-23 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of an engineered multidrug transporter MdfA reveals the molecular basis for substrate recognition.
Commun Biol, 2, 2019
|
|
7LJA
 
 | |
4AKI
 
 | | Dynein Motor Domain - LuAc derivative | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLUTATHIONE S-TRANSFERASE CLASS-MU 26 KDA ISOZYME, DYNEIN HEAVY CHAIN CYTOPLASMIC, ... | | Authors: | Schmidt, H, Gleave, E.S, Carter, A.P. | | Deposit date: | 2012-02-22 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Insights Into Dynein Motor Domain Function from a 3.3 Angstrom Crystal Structure
Nat.Struct.Mol.Biol., 19, 2012
|
|
5B6Y
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 36.2 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-06-02 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5EG1
 
 | | Antibacterial peptide ABC transporter McjD with a resolved lipid | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, MAGNESIUM ION, Microcin-J25 export ATP-binding/permease protein McjD, ... | | Authors: | Choudhury, H.G, Beis, K. | | Deposit date: | 2015-10-26 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structural and Functional Basis for Lipid Synergy on the Activity of the Antibacterial Peptide ABC Transporter McjD.
J.Biol.Chem., 291, 2016
|
|
7Q1P
 
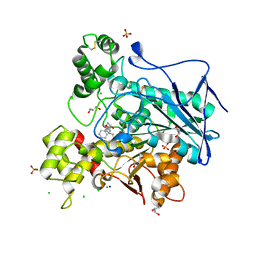 | | Crystal structure of human butyrylcholinesterase in complex with N-[(2R)-3-[(cyclohexylmethyl)amino]-2-hydroxypropyl]-3,3-diphenylpropanamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Brazzolotto, X, Panek, D, Pasieka, A, Malawska, B, Nachon, F. | | Deposit date: | 2021-10-20 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of new, highly potent and selective inhibitors of BuChE - design, synthesis, in vitro and in vivo evaluation and crystallography studies.
Eur.J.Med.Chem., 249, 2023
|
|
7LJ5
 
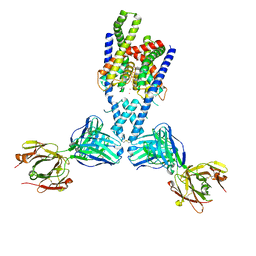 | |
7LJB
 
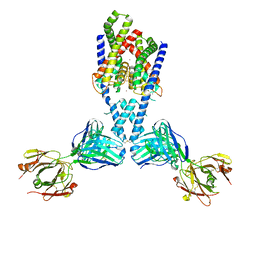 | |
7LJ4
 
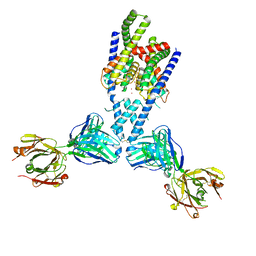 | |
4HCC
 
 | | The zinc ion bound form of crystal structure of E.coli ExoI-ssDNA complex | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), Exodeoxyribonuclease I, ISOPROPYL ALCOHOL, ... | | Authors: | Qiu, R, Wei, J, Lou, T, Liu, M, Ji, C, Gong, W. | | Deposit date: | 2012-09-29 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The structures of Escherichia coli exonuclease I in complex with the single strand DNA
To be published
|
|
6OQN
 
 | | Crystal structure of Mcl1 with inhibitor 7 | | Descriptor: | (4S)-5'-chloro-2',3',7,8,9,10,11,12-octahydro-3H,5H,14H-spiro[1,19-etheno-16lambda~6~-[1,4]oxazepino[3,4-i][1,4,5,10]oxathiadiazacyclohexadecine-4,1'-indene]-16,16,18(15H,17H)-trione, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X. | | Deposit date: | 2019-04-26 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | AMG 176, a Selective MCL1 Inhibitor, Is Effective in Hematologic Cancer Models Alone and in Combination with Established Therapies.
Cancer Discov, 8, 2018
|
|
6ZI5
 
 | | Ultrafast Structural Response to Charge Redistribution Within a Photosynthetic Reaction Centre - 300 ps (a) structure | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Baath, P, Dods, R, Braenden, G, Neutze, R. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ultrafast structural changes within a photosynthetic reaction centre.
Nature, 589, 2021
|
|
6OQU
 
 | | E. coli ATP synthase State 1d | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
4AC5
 
 | | Lipidic sponge phase crystal structure of the Bl. viridis reaction centre solved using serial femtosecond crystallography | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Johansson, L.C, Arnlund, D, White, T.A, Katona, G, DePonte, D.P, Weierstall, U, Doak, R.B, Shoeman, R.L, Lomb, L, Malmerberg, E, Davidsson, J, Nass, K, Liang, M, Andreasson, J, Aquila, A, Bajt, S, Barthelmess, M, Barty, A, Bogan, M.J, Bostedt, C, Bozek, J.D, Caleman, C, Coffee, R, Coppola, N, Ekeberg, T, Epp, S.W, Erk, B, Fleckenstein, H, Foucar, L, Graafsma, H, Gumprecht, L, Hajdu, J, Hampton, C.Y, Hartmann, R, Hartmann, A, Hauser, G, Hirsemann, H, Holl, P, Hunter, M.S, Kassemeyer, S, Kimmel, N, Kirian, R.A, Maia, F.R.N.C, Marchesini, S, Martin, A.V, Reich, C, Rolles, D, Rudek, B, Rudenko, A, Schlichting, I, Schulz, J, Seibert, M.M, Sierra, R, Soltau, H, Starodub, D, Stellato, F, Stern, S, Struder, L, Timneanu, N, Ullrich, J, Wahlgren, W.Y, Wang, X, Weidenspointner, G, Wunderer, C, Fromme, P, Chapman, H.N, Spence, J.C.H, Neutze, R. | | Deposit date: | 2011-12-14 | | Release date: | 2012-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (8.2 Å) | | Cite: | Lipidic Phase Membrane Protein Serial Femtosecond Crystallography.
Nat.Methods, 9, 2012
|
|
4HAQ
 
 | | Crystal Structure of a GH7 family cellobiohydrolase from Limnoria quadripunctata in complex with cellobiose and cellotriose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GH7 family protein, ... | | Authors: | Martin, R.N.A, McGeehan, J.E, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2012-09-27 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
