5IVN
 
 | | BC2 nanobody in complex with the BC2 peptide tag | | Descriptor: | BC2-nanobody, Cadherin derived peptide | | Authors: | Braun, M.B, Stehle, T. | | Deposit date: | 2016-03-21 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Peptides in headlock - a novel high-affinity and versatile peptide-binding nanobody for proteomics and microscopy.
Sci Rep, 6, 2016
|
|
7BR5
 
 | | Lysozyme-sugar complex in H2O | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Tanaka, I, Chatake, T. | | Deposit date: | 2020-03-26 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Recent structural insights into the mechanism of lysozyme hydrolysis.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6Y0H
 
 | | High resolution structure of GH11 xylanase from Nectria haematococca | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Andaleeb, H, Betzel, C, Perbandt, M, Brognaro, H. | | Deposit date: | 2020-02-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structure and biochemical characterization of a GH11 endoxylanase from Nectria haematococca.
Sci Rep, 10, 2020
|
|
6RZX
 
 | | Carbonic Anhydrase CAIX mimic in complex with inhibitor FBSA | | Descriptor: | 1,1,2,2,3,3,4,4,4-nonakis(fluoranyl)butane-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Kugler, M, Brynda, J, Rezacova, P. | | Deposit date: | 2019-06-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Identification of Novel Carbonic Anhydrase IX Inhibitors Using High-Throughput Screening of Pooled Compound Libraries by DNA-Linked Inhibitor Antibody Assay (DIANA).
SLAS Discov, 25, 2020
|
|
3C1P
 
 | | Crystal Structure of an alternating D-Alanyl, L-Homoalanyl PNA | | Descriptor: | Peptide Nucleic Acid DLY-HGL-AGD-LHC-AGD-LHC-CUD-LYS | | Authors: | Cuesta-Seijo, J.A, Sheldrick, G.M, Zhang, J, Diederichsen, U. | | Deposit date: | 2008-01-23 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Continuous beta-turn fold of an alternating alanyl/homoalanyl peptide nucleic acid.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4N1I
 
 | | Crystal Structure of the alpha-L-arabinofuranosidase UmAbf62A from Ustilago maidys | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Siguier, B, Dumon, C, Mourey, L, Tranier, S. | | Deposit date: | 2013-10-04 | | Release date: | 2014-01-15 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | First Structural Insights into alpha-L-Arabinofuranosidases from the Two GH62 Glycoside Hydrolase Subfamilies.
J.Biol.Chem., 289, 2014
|
|
6CDX
 
 | | High-resolution crystal structure of fluoropropylated cystine knot, binding to alpha-5 beta-6 integrin | | Descriptor: | cystine knot (fluoropropylated) | | Authors: | Kimura, R, Nix, J, Bongura, C, Chakraborti, S, Gambhir, S, Filipp, F.V. | | Deposit date: | 2018-02-09 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Evaluation of integrin alpha v beta6cystine knot PET tracers to detect cancer and idiopathic pulmonary fibrosis.
Nat Commun, 10, 2019
|
|
2WYT
 
 | | 1.0 A resolution structure of L38V SOD1 mutant | | Descriptor: | ACETATE ION, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2009-11-20 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural Discovery of Small Molecule Binding Sites in Cu-Zn Human Superoxide Dismutase Familial Amyotrophic Lateral Sclerosis Mutants Provides Insights for Lead Optimization.
J.Med.Chem., 53, 2010
|
|
2PL7
 
 | |
6YZU
 
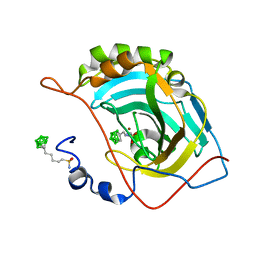 | | Carborane nido-pentyl-sulfonamide in complex with CA II | | Descriptor: | Carbonic anhydrase 2, Carborane nido-pentyl-sulfonamide, ZINC ION | | Authors: | Kugler, M, Brynda, J, Pospisilova, K, Rezacova, P. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The structural basis for the selectivity of sulfonamido dicarbaboranes toward cancer-associated carbonic anhydrase IX.
J Enzyme Inhib Med Chem, 35, 2020
|
|
5XQU
 
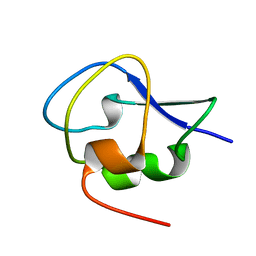 | | Crystal structure of Notched-fin eelpout type III antifreeze protein A20I mutant (NFE6, AFP), P212121 form | | Descriptor: | Ice-structuring protein | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kondo, H, Tsuda, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1LKK
 
 | |
8JE9
 
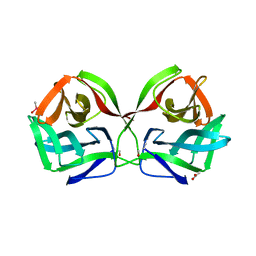 | | Crystal structure of CGL1 from Crassostrea gigas, mannobiose-bound form (CGL1/Man(alpha)1-2Man) | | Descriptor: | ACETIC ACID, CACODYLATE ION, Natterin-3, ... | | Authors: | Unno, H, Hatakeyama, T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Mannose oligosaccharide recognition of CGL1, a mannose-specific lectin containing DM9 motifs from Crassostrea gigas, revealed by X-ray crystallographic analysis.
J.Biochem., 175, 2023
|
|
6T2L
 
 | | Streptavidin variants harbouring an artificial organocatalyst based cofactor | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-(1-pyridin-4-ylpiperidin-4-yl)pentanamide, GLYCEROL, ... | | Authors: | Lechner, H, Hocker, B. | | Deposit date: | 2019-10-09 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | An Artificial Cofactor Catalyzing the Baylis-Hillman Reaction with Designed Streptavidin as Protein Host*.
Chembiochem, 22, 2021
|
|
3E7R
 
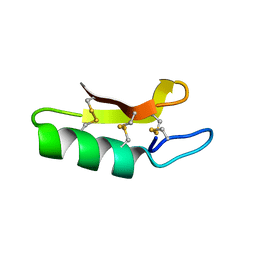 | | X-ray Crystal Structure of Racemic Plectasin | | Descriptor: | Plectasin | | Authors: | Mandal, K, Pentelute, B.L, Tereshko, V, Kossiakoff, A.A, Kent, S.B.H. | | Deposit date: | 2008-08-18 | | Release date: | 2009-06-09 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Racemic crystallography of synthetic protein enantiomers used to determine the X-ray structure of plectasin by direct methods
Protein Sci., 18, 2009
|
|
7UCR
 
 | | Joint X-ray/neutron structure of the Sarcin-Ricin loop RNA | | Descriptor: | SULFATE ION, Sarcin-Ricin loop RNA | | Authors: | Harp, J.M, Egli, M.E, Pallan, P.S, Coates, L. | | Deposit date: | 2022-03-17 | | Release date: | 2022-07-20 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1 Å), X-RAY DIFFRACTION | | Cite: | Cryo neutron crystallography demonstrates influence of RNA 2'-OH orientation on conformation, sugar pucker and water structure.
Nucleic Acids Res., 50, 2022
|
|
5R43
 
 | | Crystal Structure of deuterated gamma-Chymotrypsin at pH 7.5, cryo temperature | | Descriptor: | Chymotrypsinogen A, IODIDE ION, MALONIC ACID, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5XQP
 
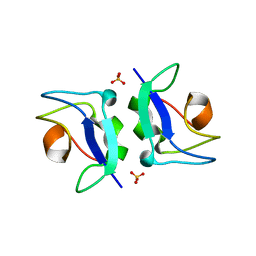 | | Crystal structure of Notched-fin eelpout type III antifreeze protein (NFE6, AFP), P212121 form | | Descriptor: | Ice-structuring protein, SULFATE ION | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kondo, H, Tsuda, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3P8J
 
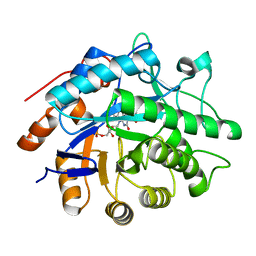 | |
4YNH
 
 | |
2AT8
 
 | |
1R2M
 
 | | Atomic resolution structure of the HFBII hydrophobin: a self-assembling amphiphile | | Descriptor: | Hydrophobin II, MANGANESE (II) ION | | Authors: | Hakanpaa, J, Paananen, A, Askolin, S, Nakari-Setala, T, Parkkinen, T, Penttila, M, Linder, M.B, Rouvinen, J. | | Deposit date: | 2003-09-29 | | Release date: | 2004-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution structure of the HFBII hydrophobin, a self-assembling amphiphile.
J.Biol.Chem., 279, 2004
|
|
4DP9
 
 | | The 1.00 Angstrom crystal structure of oxidized (CuII) poplar plastocyanin A at pH 6.0 | | Descriptor: | COPPER (II) ION, Plastocyanin A, chloroplastic | | Authors: | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
5LXW
 
 | |
4DRQ
 
 | | Exploration of Pipecolate Sulfonamides as Binders of the FK506-Binding Proteins 51 and 52: Complex of FKBP51 with 2-(3-((R)-1-((S)-1-(3,5-dichlorophenylsulfonyl)piperidine-2-carbonyloxy)-3-(3,4-dimethoxy -phenyl)propyl)phenoxy)acetic acid | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1S)-1-[({(2S)-1-[(3,5-dichlorophenyl)sulfonyl]piperidin-2-yl}carbonyl)oxy]-3-(3,4-dimethoxyphenyl)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Wang, Y, Hoogeland, B, Bracher, A, Hausch, F, Schneider, S. | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Exploration of Pipecolate Sulfonamides as Binders of the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
