8Q0F
 
 | | Inward-facing, open2 proteoliposome complex I at 3.1 A. Initially purified in DDM. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Grba, D.N, Hirst, J. | | Deposit date: | 2023-07-28 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular mechanism of the ischemia-induced regulatory switch in mammalian complex I.
Science, 384, 2024
|
|
7N17
 
 | | Structure of TAX-4_R421W apo open state | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel | | Authors: | Zheng, X, Li, H, Hu, Z, Su, D, Yang, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel.
Commun Biol, 5, 2022
|
|
5H2N
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 95.2 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
4A5W
 
 | | Crystal structure of C5b6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COMPLEMENT C5, ... | | Authors: | Hadders, M.A, Bubeck, D, Forneris, F, Pangburn, M, Llorca, O, Lea, S.M, Gros, P. | | Deposit date: | 2011-10-28 | | Release date: | 2012-03-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Assembly and Regulation of the Membrane Attack Complex Based on Structures of C5B6 and Sc5B9.
Cell Rep., 1, 2012
|
|
4GJY
 
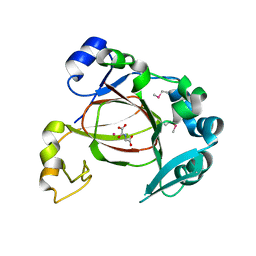 | | JMJD5 in complex with N-Oxalylglycine | | Descriptor: | COBALT (II) ION, JmjC domain-containing protein 5, N-OXALYLGLYCINE | | Authors: | Del Rizzo, P.A, Trievel, R.C. | | Deposit date: | 2012-08-10 | | Release date: | 2012-09-05 | | Last modified: | 2012-11-14 | | Method: | X-RAY DIFFRACTION (1.2492 Å) | | Cite: | Crystal Structure and Functional Analysis of JMJD5 Indicate an Alternate Specificity and Function.
Mol.Cell.Biol., 32, 2012
|
|
4A7Y
 
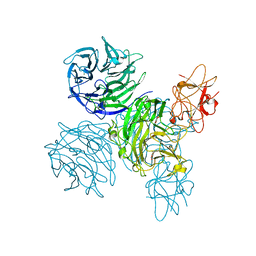 | | Active site metal depleted aldos-2-ulose dehydratase | | Descriptor: | 1,5-anhydro-D-fructose, ALDOS-2-ULOSE DEHYDRATASE, MAGNESIUM ION, ... | | Authors: | Claesson, M, Lindqvist, Y, Madrid, S, Sandalova, T, Fiskesund, R, Yu, S, Schneider, G. | | Deposit date: | 2011-11-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Bifunctional Aldos-2-Ulose Dehydratase/Isomerase from Phanerochaete Chrysosporium with the Reaction Intermediate Ascopyrone M.
J.Mol.Biol., 417, 2012
|
|
7NAK
 
 | | Cryo-EM structure of activated human SARM1 in complex with NMN and 1AD (TIR:1AD) | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(5-iodanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Kerry, P.S, Nanson, J.D, Adams, S, Cunnea, K, Bosanac, T, Kobe, B, Hughes, R.O, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
7MUB
 
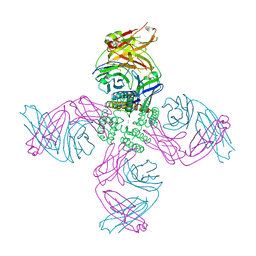 | | KcsA Open gate E71V mutant in Potassium | | Descriptor: | Fab heavy chain, Fab light chain, POTASSIUM ION, ... | | Authors: | Rohaim, A, Li, J, Weingarth, M, Roux, B. | | Deposit date: | 2021-05-14 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A distinct mechanism of C-type inactivation in the Kv-like KcsA mutant E71V.
Nat Commun, 13, 2022
|
|
5A1A
 
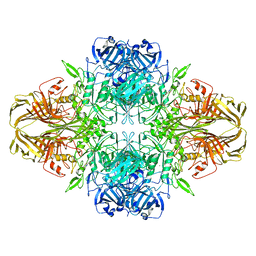 | | 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor | | Descriptor: | 2-phenylethyl 1-thio-beta-D-galactopyranoside, BETA-GALACTOSIDASE, MAGNESIUM ION, ... | | Authors: | Bartesaghi, A, Merk, A, Banerjee, S, Matthies, D, Wu, X, Milne, J, Subramaniam, S. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with a Cell-Permeant Inhibitor
Science, 348, 2015
|
|
6O5H
 
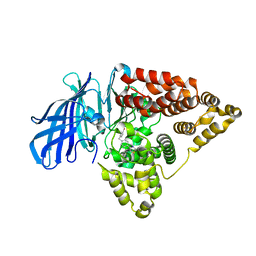 | | The effect of modifier structure on the activation of leukotriene A4 hydrolase aminopeptidase activity. | | Descriptor: | 4-{4-[(4-methoxyphenyl)methyl]phenyl}-1,3-thiazol-2-amine, Leukotriene A-4 hydrolase, ZINC ION | | Authors: | Noble, S.M, Lee, K.H, Paige, M. | | Deposit date: | 2019-03-03 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Effect of Modifier Structure on the Activation of Leukotriene A4Hydrolase Aminopeptidase Activity.
J.Med.Chem., 62, 2019
|
|
3OJW
 
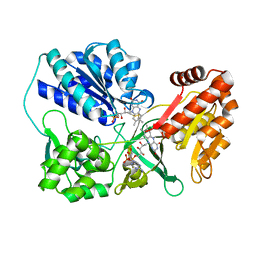 | | Disulfide crosslinked cytochrome P450 reductase inactive | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH-cytochrome P450 reductase | | Authors: | Xia, C, Hamdane, D, Shen, A, Choi, V, Kasper, C, Zhang, H, Im, S.-C, Waskell, L, Kim, J.-J.P. | | Deposit date: | 2010-08-23 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational Changes of NADPH-Cytochrome P450 Oxidoreductase Are Essential for Catalysis and Cofactor Binding.
J.Biol.Chem., 286, 2011
|
|
6OEV
 
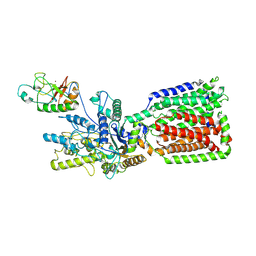 | | Structure of human Patched1 in complex with native Sonic Hedgehog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein patched homolog 1, ... | | Authors: | Qi, X, Li, X. | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-17 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of human Patched and its complex with native palmitoylated sonic hedgehog.
Nature, 560, 2018
|
|
5GTH
 
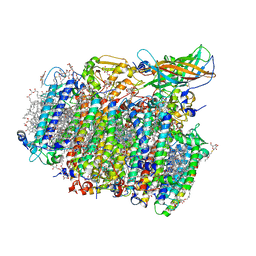 | | Native XFEL structure of photosystem II (dark dataset) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2016-08-20 | | Release date: | 2017-03-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Light-induced structural changes and the site of O=O bond formation in PSII caught by XFEL.
Nature, 543, 2017
|
|
5A3H
 
 | | 2-DEOXY-2-FLURO-B-D-CELLOBIOSYL/ENZYME INTERMEDIATE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | ENDOGLUCANASE, beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Davies, G.J, Varrot, A, Dauter, M, Brzozowski, A.M, Schulein, M, Mackenzie, L, Withers, S.G. | | Deposit date: | 1998-07-23 | | Release date: | 1999-07-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase.
Biochemistry, 37, 1998
|
|
8QE8
 
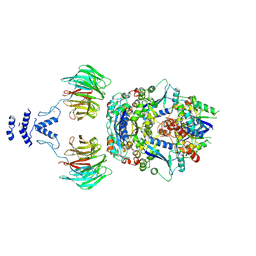 | | Structure of the non-canonical CTLH E3 substrate receptor WDR26 bound to NMNAT1 substrate | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Nicotinamide/nicotinic acid mononucleotide adenylyltransferase 1, WD repeat-containing protein 26, ... | | Authors: | Chrustowicz, J, Sherpa, D, Schulman, B.A. | | Deposit date: | 2023-08-30 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Non-canonical substrate recognition by the human WDR26-CTLH E3 ligase regulates prodrug metabolism.
Mol.Cell, 84, 2024
|
|
6O87
 
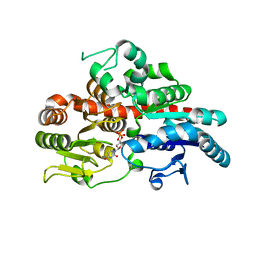 | |
5G36
 
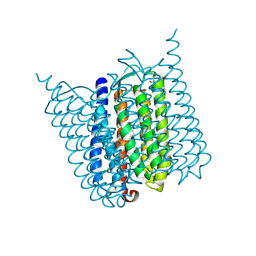 | | Yellow form of Halorhodopsin from Halobacterium salinarum in a new rhombohedral crystal form | | Descriptor: | CHLORIDE ION, HALORHODOPSIN, RETINAL, ... | | Authors: | Schreiner, M, Schlesinger, R, Heberle, J, Niemann, H.H. | | Deposit date: | 2016-04-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Halobacterium Salinarum Halorhodopsin with Partially Depopulated Primary Chloride Binding Site
Acta Crystallogr.,Sect.F, 72, 2016
|
|
7B0N
 
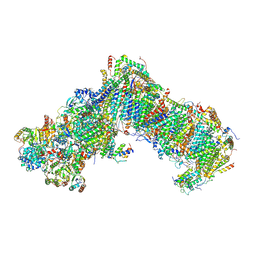 | |
7NAL
 
 | | Cryo-EM structure of activated human SARM1 in complex with NMN and 1AD (ARM and SAM domains) | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1 | | Authors: | Kerry, P.S, Nanson, J.D, Adams, S, Cunnea, K, Bosanac, T, Kobe, B, Hughes, R.O, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
8Q0Q
 
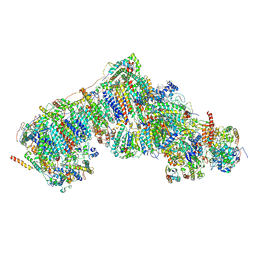 | | Outward-facing, slack proteoliposome complex I at 3.6 A. Initially purified in DDM | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Grba, D.N, Hirst, J. | | Deposit date: | 2023-07-28 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular mechanism of the ischemia-induced regulatory switch in mammalian complex I.
Science, 384, 2024
|
|
7AHI
 
 | | Substrate-engaged type 3 secretion system needle complex from Salmonella enterica typhimurium - SpaR state 2 | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, LAURYL DIMETHYLAMINE-N-OXIDE, Lipoprotein PrgK, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Miletic, S, Wald, J, Marlovits, T. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Substrate-engaged type III secretion system structures reveal gating mechanism for unfolded protein translocation
Nat Commun, 12, 2021
|
|
4GOL
 
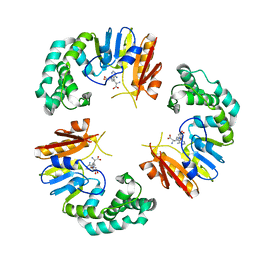 | |
4AE1
 
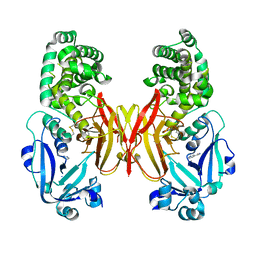 | |
4GP4
 
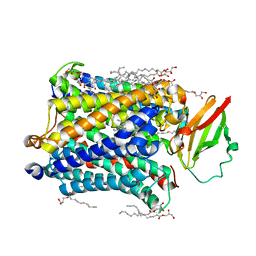 | | Structure of Recombinant Cytochrome ba3 Oxidase mutant Y133F from Thermus thermophilus | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Li, Y, Chen, Y, Stout, C.D. | | Deposit date: | 2012-08-20 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ligand Access to the Active Site in Thermus thermophilusba(3) and Bovine Heart aa(3) Cytochrome Oxidases.
Biochemistry, 52, 2013
|
|
8Q2P
 
 | | Structure of the membrane integral lipoprotein N-acyltransferase Lnt from E. coli by using Se-MAG for the the lipid cubic phase crystallization | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Apolipoprotein N-acyltransferase, ... | | Authors: | Huang, C.-Y, Boland, C, Kaki, S.S, Wang, M, Olieric, V, Caffrey, M. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Se-MAG Is a Convenient Additive for Experimental Phasing and Structure Determination of Membrane Proteins Crystallised by the Lipid Cubic Phase (In Meso) Method
Crystals, 2023
|
|
