6PPN
 
 | | Structure of S. pombe Lsm2-8 with unprocessed U6 snRNA | | Descriptor: | Mimic of unprocessed U6 snRNA, Probable U6 snRNA-associated Sm-like protein LSm3, Probable U6 snRNA-associated Sm-like protein LSm4, ... | | Authors: | Montemayor, E.J, Butcher, S.E. | | Deposit date: | 2019-07-08 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Molecular basis for the distinct cellular functions of the Lsm1-7 and Lsm2-8 complexes.
Rna, 26, 2020
|
|
6PPQ
 
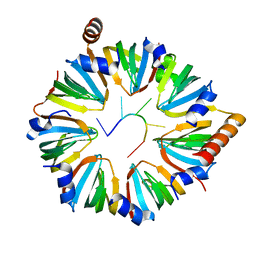 | |
6PPV
 
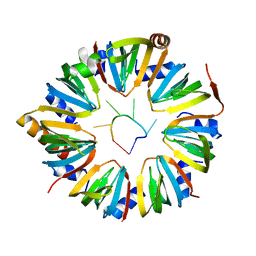 | |
6PPP
 
 | | Structure of S. pombe Lsm2-8 with processed U6 snRNA | | Descriptor: | Mimic of processed U6 snRNA, Probable U6 snRNA-associated Sm-like protein LSm3, Probable U6 snRNA-associated Sm-like protein LSm4, ... | | Authors: | Montemayor, E.J, Butcher, S.E. | | Deposit date: | 2019-07-08 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Molecular basis for the distinct cellular functions of the Lsm1-7 and Lsm2-8 complexes.
Rna, 26, 2020
|
|
7B7D
 
 | | Yeast 80S ribosome bound to eEF3 and A/A- and P/P-tRNAs | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Ranjan, N, Pochopien, A.A, Wu, C.C, Beckert, B, Blanchet, S, Green, R, Rodnina, M.V, Wilson, D.N. | | Deposit date: | 2020-12-10 | | Release date: | 2021-03-10 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | eEF3 promotes late stages of tRNA translocation including E-tRNA release from the ribosome
To Be Published
|
|
6AH3
 
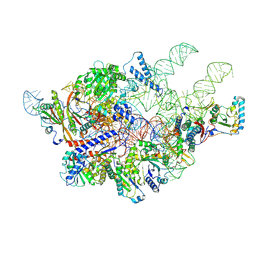 | | Cryo-EM structure of yeast Ribonuclease P with pre-tRNA substrate | | Descriptor: | MAGNESIUM ION, RNases MRP/P 32.9 kDa subunit, Ribonuclease P RNA, ... | | Authors: | Lan, P, Tan, M, Wu, J, Lei, M. | | Deposit date: | 2018-08-16 | | Release date: | 2018-10-17 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural insight into precursor tRNA processing by yeast ribonuclease P.
Science, 362, 2018
|
|
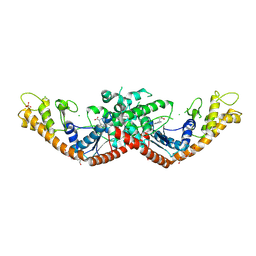
9O68
 
 | |
9LOT
 
 | | Crystal structure of Escherichia coli trptophanyl-tRNA synthetase in complex with N-piperidine ibrutinib | | Descriptor: | 3-(4-phenoxyphenyl)-1-piperidin-4-yl-pyrazolo[3,4-d]pyrimidin-4-amine, CHLORZOXAZONE, SULFATE ION, ... | | Authors: | Peng, X, Chen, B, Xia, K, Zhou, H. | | Deposit date: | 2025-01-23 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The lineage-specific tRNA recognition mechanism of bacterial trptophanyl-tRNA synthetase and its implications for inhibitor discover
To Be Published
|
|
9LPD
 
 | | Crystal structure of Escherichia coli trptophanyl-tRNA synthetase in complex with tirabrutinib | | Descriptor: | CHLORZOXAZONE, SULFATE ION, TRYPTOPHANYL-5'AMP, ... | | Authors: | Peng, X, Chen, B, Xia, K, Zhou, H. | | Deposit date: | 2025-01-24 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The lineage-specific tRNA recognition mechanism of bacterial trptophanyl-tRNA synthetase and its implications for inhibitor discover
To Be Published
|
|
9O66
 
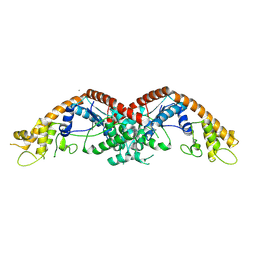 | |
9M5N
 
 | | Plasmodium vivax aspartyl-tRNA synthetase in complex with aspartyl-adenylate (Asp-AMP) Complex. | | Descriptor: | ACETATE ION, ASPARTYL-ADENOSINE-5'-MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Sharma, V.K, Manickam, Y, Sharma, A, Bagale, S, Pradeepkumar, P.I. | | Deposit date: | 2025-03-06 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Natural product-mediated reaction hijacking mechanism validates Plasmodium aspartyl-tRNA synthetase as an antimalarial drug target
To Be Published
|
|
9M5O
 
 | | Plasmodium vivax aspartyl-tRNA synthetase in complex with aspartyl sulfamoyl adenosine (Asp-AMS) Complex | | Descriptor: | 5'-O-(L-alpha-aspartylsulfamoyl)adenosine, MAGNESIUM ION, aspartate--tRNA ligase | | Authors: | Sharma, V.K, Manickam, Y, Sharma, A, Bagale, S, Pradeepkumar, P.I. | | Deposit date: | 2025-03-06 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | Natural product-mediated reaction hijacking mechanism validates Plasmodium aspartyl-tRNA synthetase as an antimalarial drug target
To Be Published
|
|
5LMU
 
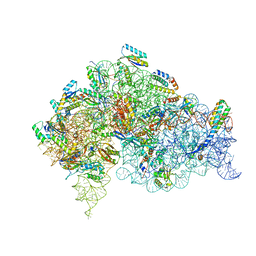 | | Structure of bacterial 30S-IF3-mRNA-tRNA translation pre-initiation complex, closed form (state-4) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
9M5M
 
 | | Crystal structure of Plasmodium vivax aspartyl-tRNA synthetase (PvDRS) | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, aspartate--tRNA ligase | | Authors: | Sharma, V.K, Manickam, Y, Bagale, S, Pradeepkumar, P.I, Sharma, A. | | Deposit date: | 2025-03-06 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Natural product-mediated reaction hijacking mechanism validates Plasmodium aspartyl-tRNA synthetase as an antimalarial drug target
To Be Published
|
|
5LMV
 
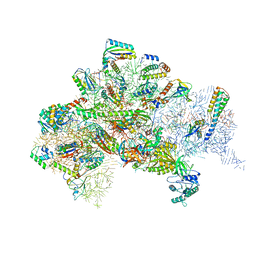 | | Structure of bacterial 30S-IF1-IF2-IF3-mRNA-tRNA translation pre-initiation complex(state-III) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
5LMT
 
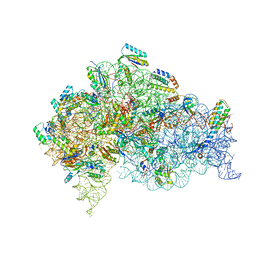 | | Structure of bacterial 30S-IF1-IF3-mRNA-tRNA translation pre-initiation complex(state-3) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
5LMR
 
 | | Structure of bacterial 30S-IF1-IF3-mRNA-tRNA translation pre-initiation complex(state-2B) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (4.45 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
5LMS
 
 | | Structure of bacterial 30S-IF1-IF3-mRNA-tRNA translation pre-initiation complex(state-2C) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
5LMQ
 
 | | Structure of bacterial 30S-IF1-IF3-mRNA-tRNA translation pre-initiation complex, open form (state-2A) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
1LS2
 
 | | Fitting of EF-Tu and tRNA in the Low Resolution Cryo-EM Map of an EF-Tu Ternary Complex (GDP and Kirromycin) Bound to E. coli 70S Ribosome | | Descriptor: | Elongation Factor Tu, Phenylalanine transfer RNA | | Authors: | Valle, M, Sengupta, J, Swami, N.K, Grassucci, R.A, Burkhardt, N, Nierhaus, K.H, Agrawal, R.K, Frank, J. | | Deposit date: | 2002-05-16 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (16.799999 Å) | | Cite: | Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process.
EMBO J., 21, 2002
|
|
4OO1
 
 | |
5ZEB
 
 | | M. Smegmatis P/P state 70S ribosome structure | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Mishra, S, Ahmed, T, Tyagi, A, Shi, J, Bhushan, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-09-26 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Mycobacterium smegmatis 70S ribosomes in complex with HPF, tmRNA, and P-tRNA.
Sci Rep, 8, 2018
|
|
6SPG
 
 | | Pseudomonas aeruginosa 70s ribosome from a clinical isolate | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-16 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5ZEU
 
 | | M. smegmatis P/P state 30S ribosomal subunit | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Mishra, S, Ahmed, T, Tyagi, A, Shi, J, Bhushan, S. | | Deposit date: | 2018-02-28 | | Release date: | 2018-09-26 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of Mycobacterium smegmatis 70S ribosomes in complex with HPF, tmRNA, and P-tRNA.
Sci Rep, 8, 2018
|
|
6PMO
 
 | |
