6GMH
 
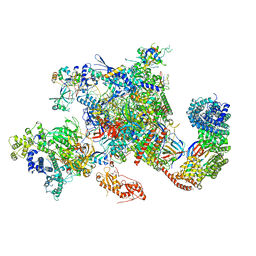 | | Structure of activated transcription complex Pol II-DSIF-PAF-SPT6 | | Descriptor: | CDC73, CTR9,RNA polymerase-associated protein CTR9 homolog,RNA polymerase-associated protein CTR9 homolog, DNA-directed RNA polymerase II subunit RPB9, ... | | Authors: | Vos, S.M, Farnung, L, Boehing, M, Linden, A, Wigge, C, Urlaub, H, Cramer, P. | | Deposit date: | 2018-05-26 | | Release date: | 2018-08-22 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of activated transcription complex Pol II-DSIF-PAF-SPT6.
Nature, 560, 2018
|
|
6W77
 
 | | 30S-Inactivated-high-Mg2+ Class A | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Jahagirdar, D, Jha, V, Basu, B, Gomez-Blanco, J, Vargas, J, Ortega, J. | | Deposit date: | 2020-03-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy.
Rna, 26, 2020
|
|
5FBC
 
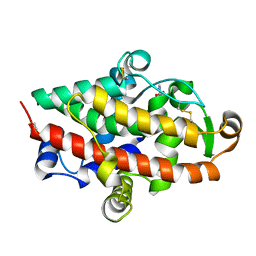 | | S1 nuclease from Aspergillus oryzae in complex with 2'-deoxyadenosine-5'-thio-monophosphate (5'dAMP(S)). | | Descriptor: | 2-DEOXY-ADENOSINE -5'-THIO-MONOPHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Catalytic Properties of S1 Nuclease from Aspergillus oryzae Responsible for Substrate Recognition, Cleavage, Non-Specificity, and Inhibition.
PLoS ONE, 11, 2016
|
|
5F9L
 
 | |
7XFG
 
 | |
6W7M
 
 | | 30S-Inactive-high-Mg2+ + carbon layer | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Jahagirdar, D, Jha, V, Basu, B, Gomez-Blanco, J, Vargas, J, Ortega, J. | | Deposit date: | 2020-03-19 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy.
Rna, 26, 2020
|
|
8VGL
 
 | | CryoEM structure of Nav1.7 in complex with wild type Fab 7A9 | | Descriptor: | Chimeric Nav1.7-NavAb, Fab 7A9 heavy chain, Fab 7A9 light chain | | Authors: | Kung, J.E, Jao, C.C, Arthur, C.P, Sudhamsu, J. | | Deposit date: | 2023-12-27 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Disulfi de constrained Fabs overcome target size limitation for high-resolution single-particle cryo-EM.
Biorxiv, 2024
|
|
6VOF
 
 | | Chloroplast ATP synthase (O2, CF1FO) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.51 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
6GY6
 
 | |
6W7W
 
 | | 30S-Inactive-low-Mg2+ Class B | | Descriptor: | 16S rRNA, 30S ribosomal protein S12, 30S ribosomal protein S15, ... | | Authors: | Jahagirdar, D, Jha, V, Basu, B, Gomez-Blanco, J, Vargas, J, Ortega, J. | | Deposit date: | 2020-03-19 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy.
Rna, 26, 2020
|
|
6VM4
 
 | | Chloroplast ATP synthase (C2, CF1FO) | | Descriptor: | ATP synthase delta chain, chloroplastic, ATP synthase epsilon chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.08 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
6VON
 
 | | Chloroplast ATP synthase (R1, CF1FO) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
3M6M
 
 | | Crystal structure of RpfF complexed with REC domain of RpfC | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Cheng, Z, Lim, S.C, Qamra, R, Song, H. | | Deposit date: | 2010-03-16 | | Release date: | 2010-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of the Sensor-Synthase Interaction in Autoinduction of the Quorum Sensing Signal DSF Biosynthesis
Structure, 18, 2010
|
|
5XUJ
 
 | | Crystal structure of PDE10A in complex with 7-(4-chlorophenyl)-2-methylpyrazolo[1,5-a]pyrimidine | | Descriptor: | 7-(4-chlorophenyl)-2-methyl-pyrazolo[1,5-a]pyrimidine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Amano, Y, Honbou, K. | | Deposit date: | 2017-06-23 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Fragment-Based Discovery of Pyrimido[1,2-b]indazole PDE10A Inhibitors.
Chem. Pharm. Bull., 66, 2018
|
|
6VOL
 
 | | Chloroplast ATP synthase (R2, CF1FO) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
6VOH
 
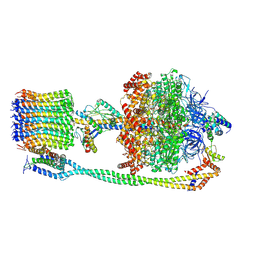 | | Chloroplast ATP synthase (O1, CF1FO) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-30 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
6M6A
 
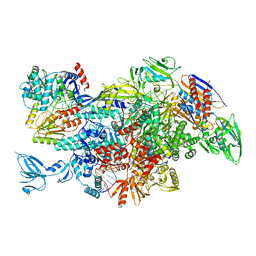 | | Cryo-EM structure of Thermus thermophilus Mfd in complex with RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
5GAS
 
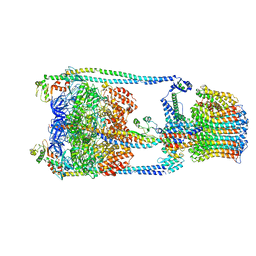 | | Thermus thermophilus V/A-ATPase, conformation 2 | | Descriptor: | Archaeal/vacuolar-type H+-ATPase subunit I, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Schep, D.G, Zhao, J, Rubinstein, J.L. | | Deposit date: | 2016-02-05 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Models for the a subunits of the Thermus thermophilus V/A-ATPase and Saccharomyces cerevisiae V-ATPase enzymes by cryo-EM and evolutionary covariance.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5I2K
 
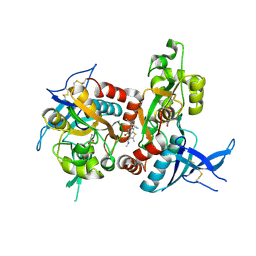 | | Structure of the human GluN1/GluN2A LBD in complex with 7-{[ethyl(4-fluorophenyl)amino]methyl}-N,2-dimethyl-5-oxo-5H-[1,3]thiazolo[3,2-a]pyrimidine-3-carboxamide (compound 19) | | Descriptor: | 7-{[ethyl(4-fluorophenyl)amino]methyl}-N,2-dimethyl-5-oxo-5H-[1,3]thiazolo[3,2-a]pyrimidine-3-carboxamide, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-02-09 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Discovery of GluN2A-Selective NMDA Receptor Positive Allosteric Modulators (PAMs): Tuning Deactivation Kinetics via Structure-Based Design.
J.Med.Chem., 59, 2016
|
|
6VOJ
 
 | | Chloroplast ATP synthase (R3, CF1FO) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
6VOM
 
 | | Chloroplast ATP synthase (R2, CF1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
5I2N
 
 | | Structure of the human GluN1/GluN2A LBD in complex with N-ethyl-7-{[2-fluoro-3-(trifluoromethyl)phenyl]methyl}-2-methyl-5-oxo-5H-[1,3]thiazolo[3,2-a]pyrimidine-3-carboxamide (compound 29) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, GLUTAMIC ACID, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-02-09 | | Release date: | 2016-03-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Discovery of GluN2A-Selective NMDA Receptor Positive Allosteric Modulators (PAMs): Tuning Deactivation Kinetics via Structure-Based Design.
J.Med.Chem., 59, 2016
|
|
6VMD
 
 | | Chloroplast ATP synthase (C1, CF1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-27 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (4.53 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
6WXB
 
 | |
5IDM
 
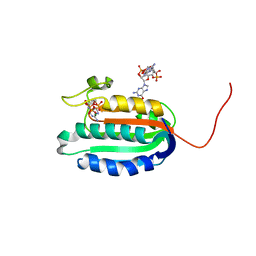 | | Bifunctional histidine kinase CckA (domain, CA) in complex with c-di-GMP and AMPPNP/Mg2+ | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Cell cycle histidine kinase CckA, MAGNESIUM ION, ... | | Authors: | Dubey, B.N, Schirmer, T. | | Deposit date: | 2016-02-24 | | Release date: | 2016-10-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cyclic di-GMP mediates a histidine kinase/phosphatase switch by noncovalent domain cross-linking.
Sci Adv, 2, 2016
|
|
