2ZQ0
 
 | | Crystal structure of SusB complexed with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase (Alpha-glucosidase SusB), CALCIUM ION | | Authors: | Yao, M, Tanaka, I, Kitamura, M. | | Deposit date: | 2008-07-31 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides thetaiotaomicron.
J.Biol.Chem., 283, 2008
|
|
2ZTZ
 
 | |
3A7O
 
 | |
2H5R
 
 | |
3C3I
 
 | | Evolution of chlorella virus dUTPase | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, Deoxyuridine triphosphatase, MAGNESIUM ION | | Authors: | Yamanishi, M, Homma, K, Zhang, Y, Etten, L.V.J, Moriyama, H. | | Deposit date: | 2008-01-28 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallization and crystal-packing studies of Chlorella virus deoxyuridine triphosphatase.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3C5F
 
 | | Structure of a binary complex of the R517A Pol lambda mutant | | Descriptor: | DNA (5'-D(*DCP*DAP*DGP*DTP*DAP*DC)-3'), DNA (5'-D(*DCP*DGP*DGP*DCP*DCP*DGP*DTP*DAP*DCP*DTP*DG)-3'), DNA (5'-D(P*DGP*DCP*DCP*DG)-3'), ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Foley, M.C, Pedersen, L.C, Schlick, T, Kunkel, T.A. | | Deposit date: | 2008-01-31 | | Release date: | 2008-09-02 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate-induced DNA strand misalignment during catalytic cycling by DNA polymerase lambda.
Embo Rep., 9, 2008
|
|
2ZJY
 
 | | Structure of the K349P mutant of Gi alpha 1 subunit bound to ALF4 and GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i), alpha-1 subunit, ... | | Authors: | Morikawa, T, Muroya, A, Sugio, S, Wakamatsu, K, Kohno, T. | | Deposit date: | 2008-03-11 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | How GPCRs activate G proteins: Structural changes form C-terminal tail to GDP binding pocket
To be Published
|
|
3C2T
 
 | | Evolution of chlorella virus dUTPase | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, Deoxyuridine triphosphatase, MAGNESIUM ION | | Authors: | Yamanishi, M, Homma, K, Zhang, Y, Etten, L.V.J, Moriyama, H. | | Deposit date: | 2008-01-25 | | Release date: | 2009-02-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallization and crystal-packing studies of Chlorella virus deoxyuridine triphosphatase.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3CCX
 
 | | ALTERING SUBSTRATE SPECIFICITY AT THE HEME EDGE OF CYTOCHROME C PEROXIDASE | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wilcox, S.K, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1995-03-17 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Altering substrate specificity at the heme edge of cytochrome c peroxidase.
Biochemistry, 35, 1996
|
|
3BM5
 
 | | Crystal structure of O-acetyl-serine sulfhydrylase from Entamoeba histolytica in complex with cysteine | | Descriptor: | CYSTEINE, Cysteine synthase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Krishna, C, Kumar, M, Kumar, S, Gourinath, S. | | Deposit date: | 2007-12-12 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of native O-acetyl-serine sulfhydrylase from Entamoeba histolytica and its complex with cysteine: structural evidence for cysteine binding and lack of interactions with serine acetyl transferase.
Proteins, 72, 2008
|
|
1H29
 
 | | Sulfate respiration in Desulfovibrio vulgaris Hildenborough: Structure of the 16-heme Cytochrome c HmcA at 2.5 A resolution and a view of its role in transmembrane electron transfer | | Descriptor: | HEME C, HIGH-MOLECULAR-WEIGHT CYTOCHROME C | | Authors: | Matias, P.M, Coelho, A.V, Valente, F.M.A, Placido, D, Legall, J, Xavier, A.V, Pereira, I.A.C, Carrondo, M.A. | | Deposit date: | 2002-08-01 | | Release date: | 2002-10-02 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Sulfate Respiration in Desulfovibrio Vulgaris Hildenborough: Structure of the 16-Heme Cytochrome C Hmca at 2.5 A Resolution and a View of its Role in Transmembrane Electron Transfer
J.Biol.Chem., 277, 2002
|
|
4OU3
 
 | | Crystal structure of porcine aminopeptidase N complexed with CNGRCG tumor-homing peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Liu, C, Yang, Y, Chen, L, Lin, Y.-L, Li, F. | | Deposit date: | 2014-02-14 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Unified Mechanism for Aminopeptidase N-based Tumor Cell Motility and Tumor-homing Therapy.
J.Biol.Chem., 289, 2014
|
|
4YHE
 
 | | NATIVE BACTEROIDETES-AFFILIATED GH5 CELLULASE LINKED WITH A POLYSACCHARIDE UTILIZATION LOCUS | | Descriptor: | GH5 | | Authors: | Naas, A.E, MacKenzie, A.K, Dalhus, B, Eijsink, V.G.H, Pope, P.B. | | Deposit date: | 2015-02-27 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Features of a Bacteroidetes-Affiliated Cellulase Linked with a Polysaccharide Utilization Locus.
Sci Rep, 5, 2015
|
|
4OUN
 
 | | Crystal Structure of Mini-ribonuclease 3 from Bacillus subtilis | | Descriptor: | Mini-ribonuclease 3 | | Authors: | Chojnowski, G, Czarnecka, J, Nowak, E, Pianka, D, Glow, D, Sabala, I, Skowronek, K, Nowotny, M, Bujnicki, J.M. | | Deposit date: | 2014-02-18 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sequence-specific cleavage of dsRNA by Mini-III RNase
Nucleic Acids Res., 43, 2015
|
|
4OP3
 
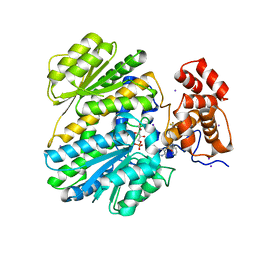 | | Human GKRP bound to AMG-5112 and Sorbitol-6-phosphate | | Descriptor: | (2S)-2-{6'-[(6-aminopyridin-3-yl)sulfonyl]-2'-(phenylamino)-2,3'-bipyridin-5-yl}-1,1,1-trifluoropropan-2-ol, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | Authors: | Jordan, S.R, Chmait, S. | | Deposit date: | 2014-02-04 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Small Molecule Disruptors of the Glucokinase-Glucokinase Regulatory Protein Interaction: 5. A Novel Aryl Sulfone Series, Optimization Through Conformational Analysis.
J.Med.Chem., 58, 2015
|
|
4OPK
 
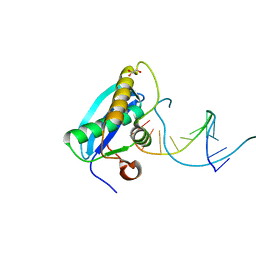 | | Bh-RNaseH:2'-SMe-DNA complex | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*(USM)P*TP*CP*GP*CP*G)-3', GLYCEROL, Ribonuclease H | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2014-02-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Generating Crystallographic Models of DNA Dodecamers from Structures of RNase H:DNA Complexes.
Methods Mol.Biol., 1320
|
|
4YIQ
 
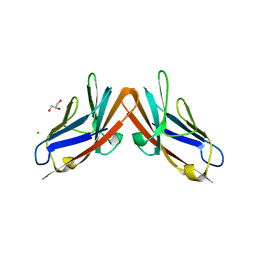 | | Structure of the CEACAM6-CEACAM8 heterodimer | | Descriptor: | CHLORIDE ION, Carcinoembryonic antigen-related cell adhesion molecule 6, Carcinoembryonic antigen-related cell adhesion molecule 8, ... | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2015-03-02 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Diverse oligomeric states of CEACAM IgV domains.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4OQI
 
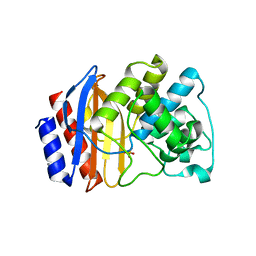 | | Crystal structure of stabilized TEM-1 beta-lactamase variant v.13 carrying R164S/G238S mutations | | Descriptor: | CALCIUM ION, SULFATE ION, TEM-94 ES-beta-lactamase | | Authors: | Dellus-Gur, E, Elias, M, Fraser, J.S, Tawfik, D.S. | | Deposit date: | 2014-02-09 | | Release date: | 2015-05-20 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Negative Epistasis and Evolvability in TEM-1 beta-Lactamase--The Thin Line between an Enzyme's Conformational Freedom and Disorder.
J. Mol. Biol., 427, 2015
|
|
4YIW
 
 | | DIHYDROOROTASE FROM BACILLUS ANTHRACIS WITH SUBSTRATE BOUND | | Descriptor: | Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ZINC ION | | Authors: | Lei, H, Santarsiero, B.D, Rice, A.J, Lee, H, Johnson, M.E. | | Deposit date: | 2015-03-02 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Ca-asp bound X-ray structure and inhibition of Bacillus anthracis dihydroorotase (DHOase).
Bioorg.Med.Chem., 24, 2016
|
|
4OQS
 
 | | Crystal structure of CYP105AS1 | | Descriptor: | CYP105AS1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Leys, D. | | Deposit date: | 2014-02-10 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Single-step fermentative production of the cholesterol-lowering drug pravastatin via reprogramming of Penicillium chrysogenum.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4YJH
 
 | | Crystal structure of DAAO(Y228L/R283G) variant (R-2-phenylpyrrolidine binding form) | | Descriptor: | (2R)-2-phenylpyrrolidine, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Nakano, S, Yasukawa, K, Kawahara, N, Ishitsubo, E, Tokiwa, H, Asano, Y. | | Deposit date: | 2015-03-03 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of DAAO variant
To Be Published
|
|
4OY9
 
 | | Crystal structure of human P-Cadherin EC1-EC2 in closed conformation | | Descriptor: | CALCIUM ION, Cadherin-3 | | Authors: | Dalle Vedove, A, Lucarelli, A.P, Nardone, V, Matino, A, Parisini, E. | | Deposit date: | 2014-02-11 | | Release date: | 2015-04-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The X-ray structure of human P-cadherin EC1-EC2 in a closed conformation provides insight into the type I cadherin dimerization pathway.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4O8N
 
 | | Crystal structure of SthAraf62A, a GH62 family alpha-L-arabinofuranosidase from Streptomyces thermoviolaceus, in the apoprotein form | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase, CALCIUM ION, ... | | Authors: | Stogios, P.J, Wang, W, Xu, X, Cui, H, Master, E, Savchenko, A. | | Deposit date: | 2013-12-28 | | Release date: | 2014-07-02 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.6476 Å) | | Cite: | Elucidation of the molecular basis for arabinoxylan-debranching activity of a thermostable family GH62 alpha-l-arabinofuranosidase from Streptomyces thermoviolaceus.
Appl.Environ.Microbiol., 80, 2014
|
|
4ORO
 
 | |
4OZ7
 
 | |
