5CCN
 
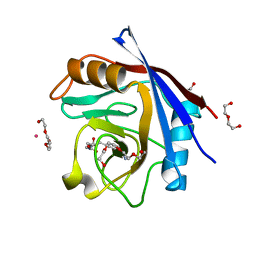 | | Human Cyclophilin D Complexed with Inhibitor | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, POTASSIUM ION, ... | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-02 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor
To Be Published
|
|
7KUT
 
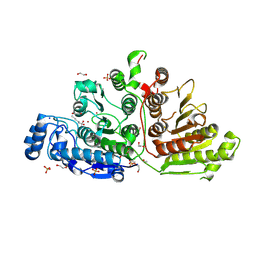 | | Crystal Structure of Danio rerio Histone Deacetylase 10 H137A Mutant in Complex with N-Acetylputrescine (Tetrahedral Intermediate) | | Descriptor: | 1,2-ETHANEDIOL, 1-[(4-aminobutyl)amino]ethane-1,1-diol, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Herbst-Gervasoni, C.J, Christianson, D.W. | | Deposit date: | 2020-11-25 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray Crystallographic Snapshots of Substrate Binding in the Active Site of Histone Deacetylase 10.
Biochemistry, 60, 2021
|
|
6F6A
 
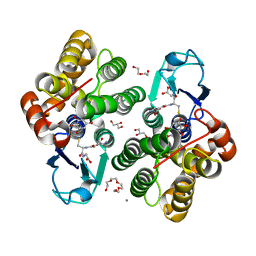 | | Crystal structure of glutathione transferase Omega 3S from Trametes versicolor in complex with dihydrowogonin from wild-cherry extract | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2017-12-05 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular recognition of wood polyphenols by phase II detoxification enzymes of the white rot Trametes versicolor.
Sci Rep, 8, 2018
|
|
7KY5
 
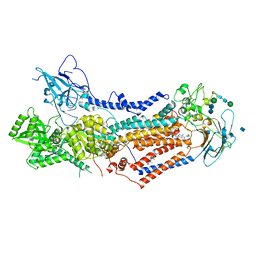 | | Structure of the S. cerevisiae phosphatidylcholine flippase Dnf2-Lem3 complex in the E2P transition state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, CHOLESTEROL, ... | | Authors: | Bai, L, You, Q, Jain, B.K, Duan, H.D, Kovach, A, Graham, T.R, Li, H. | | Deposit date: | 2020-12-07 | | Release date: | 2021-01-06 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Transport mechanism of P4 ATPase phosphatidylcholine flippases.
Elife, 9, 2020
|
|
3O5U
 
 | | Crystal Structure of 4-Chlorocatechol Dioxygenase from Rhodococcus opacus 1CP in complex with protocatechuate | | Descriptor: | (2R)-3-(PHOSPHONOOXY)-2-(TETRADECANOYLOXY)PROPYL PALMITATE, 3,4-DIHYDROXYBENZOIC ACID, CHLORIDE ION, ... | | Authors: | Ferraroni, M, Briganti, F, Kolomitseva, M, Golovleva, L. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | X-ray structures of 4-chlorocatechol 1,2-dioxygenase adducts with substituted catechols: new perspectives in the molecular basis of intradiol ring cleaving dioxygenases specificity.
J. Struct. Biol., 181, 2013
|
|
3O32
 
 | | Crystal Structure of 4-Chlorocatechol Dioxygenase from Rhodococcus opacus 1CP in complex with 3,5-dichlorocatechol | | Descriptor: | (2R)-3-(PHOSPHONOOXY)-2-(TETRADECANOYLOXY)PROPYL PALMITATE, 3,5-dichlorobenzene-1,2-diol, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Ferraroni, M, Briganti, F, Kolomytseva, M, Golovleva, L. | | Deposit date: | 2010-07-23 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | X-ray structures of 4-chlorocatechol 1,2-dioxygenase adducts with substituted catechols: new perspectives in the molecular basis of intradiol ring cleaving dioxygenases specificity.
J. Struct. Biol., 181, 2013
|
|
1F9D
 
 | | Crystal structure of the cellulase CEL48F from C. cellulolyticum in complex with cellotetraose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-GLUCANASE F, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Parsiegla, G, Reverbel-Leroy, C, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-07-10 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Cellulase Cel48F in Complex with Inhibitors and Substrates Give Insights Into its Processive Action
Biochemistry, 39, 2000
|
|
5FIP
 
 | | Discovery and characterization of a novel thermostable and highly halotolerant GH5 cellulase from an Icelandic hot spring isolate | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CHLORIDE ION, ... | | Authors: | Zarafeta, D, Kissas, D, Sayer, C, Gudbergsdottir, S.R, Ladoukakis, E, Isupov, M.N, Chatziioannou, A, Peng, X, Littlechild, J.A, Skretas, G, Kolisis, F.N. | | Deposit date: | 2015-10-01 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery and Characterization of a Thermostable and Highly Halotolerant Gh5 Cellulase from an Icelandic Hot Spring Isolate.
Plos One, 11, 2016
|
|
5CJQ
 
 | |
7KY9
 
 | | Structure of the S. cerevisiae phosphatidylcholine flippase Dnf2-Lem3 complex in the E1-ADP state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bai, L, You, Q, Jain, B.K, Duan, H.D, Kovach, A, Graham, T.R, Li, H. | | Deposit date: | 2020-12-07 | | Release date: | 2021-01-06 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Transport mechanism of P4 ATPase phosphatidylcholine flippases.
Elife, 9, 2020
|
|
7L0N
 
 | | Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Snell, G, Czudnochowski, N, Dillen, J, Nix, J.C, Croll, T.I, Corti, D. | | Deposit date: | 2020-12-11 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity.
Cell, 184, 2021
|
|
7KYB
 
 | | Structure of the S. cerevisiae phosphatidylcholine flippase Dnf1-Lem3 complex in the E1-ADP state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bai, L, You, Q, Jain, B.K, Duan, H.D, Kovach, A, Graham, T.R, Li, H. | | Deposit date: | 2020-12-07 | | Release date: | 2021-01-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Transport mechanism of P4 ATPase phosphatidylcholine flippases.
Elife, 9, 2020
|
|
7L82
 
 | | x-ray structure of the psychrobacter cryohalolentis Pcryo_0637 N-acetyltransferase in the presene of its reaction tetrahedral intermediate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Putative acetyl transferase protein, ... | | Authors: | Linehan, M.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2020-12-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterization of two enzymes from Psychrobacter cryohalolentis that are required for the biosynthesis of an unusual diacetamido-d-sugar.
J.Biol.Chem., 296, 2021
|
|
3NUF
 
 | |
3KH1
 
 | |
3NSU
 
 | | A Systematic Screen for Protein-Lipid Interactions in Saccharomyces cerevisiae | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate-binding protein SLM1, SULFATE ION | | Authors: | Gallego, O, Fernandez-Tornero, C, Aguilar-Gurrieri, C, Muller, C, Gavin, A.C. | | Deposit date: | 2010-07-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae.
Mol. Syst. Biol., 6, 2010
|
|
6F68
 
 | | Crystal structure glutathione transferase Omega 3S from Trametes versicolor in complex with 2,4,4'-trihydroxybenzophenone | | Descriptor: | (2,4-dihydroxyphenyl)(4-hydroxyphenyl)methanone, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2017-12-05 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Molecular recognition of wood polyphenols by phase II detoxification enzymes of the white rot Trametes versicolor.
Sci Rep, 8, 2018
|
|
6L0A
 
 | | Crystal structure of dihydroorotase in complex with malate at pH7 from Saccharomyces cerevisiae | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2021-12-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Analysis of Saccharomyces cerevisiae Dihydroorotase Reveals Molecular Insights into the Tetramerization Mechanism
Molecules, 2021
|
|
6EUQ
 
 | | MdfA(Q131R/L339E) | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, ACETATE ION, HEXANE, ... | | Authors: | Tam, H.K, Zomot, E, Bibi, E, Pos, K.M. | | Deposit date: | 2017-10-31 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A New Critical Conformational Determinant of Multidrug Efflux by an MFS Transporter.
J. Mol. Biol., 430, 2018
|
|
5FWD
 
 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 with CP2 | | Descriptor: | 1,2-ETHANEDIOL, 9-(7-{[amino(iminio)methyl]amino}-5,6,7-trideoxy-beta-D-ribo-heptofuranosyl)-9H-purin-6-amine, CALCIUM ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-02-12 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Studies of Protein Arginine Methyltransferase 2 Reveal its Interactions with Potential Substrates and Inhibitors.
FEBS J., 284, 2017
|
|
5CBV
 
 | | Human Cyclophilin D Complexed with Inhibitor | | Descriptor: | FORMIC ACID, Human Cyclophilin D, POTASSIUM ION, ... | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-01 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor
To Be Published
|
|
6EX7
 
 | | Crystal structure of NDM-1 metallo-beta-lactamase in complex with Cd ions and a hydrolyzed beta-lactam ligand - new refinement | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, CADMIUM ION, ... | | Authors: | Kim, Y, Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, Tesar, C, Jedrzejczak, R, Babnigg, J, Mire, J, Sacchettini, J, Joachimiak, A. | | Deposit date: | 2017-11-07 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
3O6J
 
 | | Crystal Structure of 4-Chlorocatechol Dioxygenase from Rhodococcus opacus 1CP in complex with hydroxyquinol | | Descriptor: | (2R)-3-(PHOSPHONOOXY)-2-(TETRADECANOYLOXY)PROPYL PALMITATE, CHLORIDE ION, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Ferraroni, M, Briganti, F, Kolomitseva, M, Golovleva, L. | | Deposit date: | 2010-07-29 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structures of 4-chlorocatechol 1,2-dioxygenase adducts with substituted catechols: new perspectives in the molecular basis of intradiol ring cleaving dioxygenases specificity.
J. Struct. Biol., 181, 2013
|
|
5FX5
 
 | | Novel inhibitors of human rhinovirus 3C protease | | Descriptor: | GLYCEROL, RHINOVIRUS 3C PROTEASE, TETRAETHYLENE GLYCOL, ... | | Authors: | Kawatkar, S, Ek, M, Hoesch, V, Gagnon, M, Nilsson, E, Lister, T, Olsson, L, Patel, J, Yu, Q. | | Deposit date: | 2016-02-24 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Structure-Activity Relationships of Novel Inhibitors of Human Rhinovirus 3C Protease.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5FWA
 
 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 with CP1 | | Descriptor: | 1,2-ETHANEDIOL, 9-(6-carbamimidamido-5,6-dideoxy-beta-D-ribo-hexofuranosyl)-9H-purin-6-amine, CALCIUM ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-02-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors.
FEBS J., 284, 2017
|
|
