2C2S
 
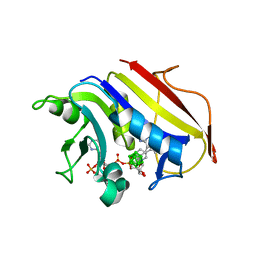 | | Human Dihydrofolate Reductase Complexed With NADPH and 2,4-Diamino-5-(1-o-carboranylmethyl)-6-methylpyrimidine, A novel boron containing, nonclassical Antifolate | | Descriptor: | 2,4-DIAMINO-5-(1-O-CARBORANYLMETHYL)-6-METHYLPYRIMIDINE, DIHYDROFOLATE REDUCTASE, GLYCEROL, ... | | Authors: | Leung, A.K.W, Reynolds, R.C, Riordan, J.M, Borhani, D.W. | | Deposit date: | 2005-09-29 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Novel Boron-Containing, Nonclassical Antifolates: Synthesis and Preliminary Biological and Structural Evaluation.
J.Med.Chem., 50, 2007
|
|
3IZ0
 
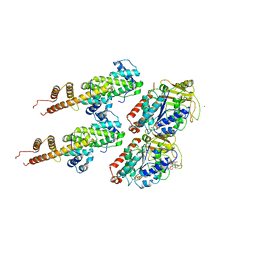 | | Human Ndc80 Bonsai Decorated Microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Alushin, G.M, Ramey, V.H, Pasqualato, S, Ball, D.A, Grigorieff, N, Musacchio, A, Nogales, E. | | Deposit date: | 2010-08-09 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | The Ndc80 kinetochore complex forms oligomeric arrays along microtubules.
Nature, 467, 2010
|
|
5T0N
 
 | |
1GK7
 
 | | HUMAN VIMENTIN COIL 1A FRAGMENT (1A) | | Descriptor: | SULFATE ION, VIMENTIN | | Authors: | Strelkov, S.V, Herrmann, H, Geisler, N, Zimbelmann, R, Aebi, U, Burkhard, P. | | Deposit date: | 2001-08-08 | | Release date: | 2002-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conserved Segments 1A and 2B of the Intermediate Filament Dimer: Their Atomic Structures and Role in Filament Assembly.
Embo J., 21, 2002
|
|
1GK4
 
 | | HUMAN VIMENTIN COIL 2B FRAGMENT (CYS2) | | Descriptor: | ACETATE ION, VIMENTIN | | Authors: | Strelkov, S.V, Herrmann, H, Geisler, N, Zimbelmann, R, Aebi, U, Burkhard, P. | | Deposit date: | 2001-08-08 | | Release date: | 2002-03-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conserved Segments 1A and 2B of the Intermediate Filament Dimer: Their Atomic Structures and Role in Filament Assembly.
Embo J., 21, 2002
|
|
4PRF
 
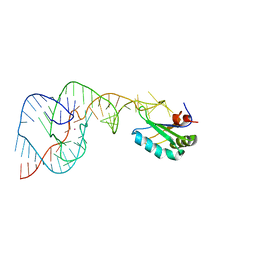 | | A Second Look at the HDV Ribozyme Structure and Dynamics. | | Descriptor: | Hepatitis Delta virus ribozyme, STRONTIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Kapral, G.J, Jain, S, Noeske, J, Doudna, J.A, Richardson, D.C, Richardson, J.S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | New tools provide a second look at HDV ribozyme structure, dynamics and cleavage.
Nucleic Acids Res., 42, 2014
|
|
1JB1
 
 | | Lactobacillus casei HprK/P Bound to Phosphate | | Descriptor: | HPRK PROTEIN, PHOSPHATE ION | | Authors: | Fieulaine, S, Morera, S, Poncet, S, Monedero, V, Gueguen-Chaignon, V, Galinier, A, Janin, J, Deutscher, J, Nessler, S. | | Deposit date: | 2001-06-01 | | Release date: | 2001-08-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of HPr kinase: a bacterial protein kinase with a P-loop nucleotide-binding domain.
EMBO J., 20, 2001
|
|
4HG7
 
 | | Crystal Structure of an MDM2/Nutlin-3a complex | | Descriptor: | 4-({(4S,5R)-4,5-bis(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Noble, M.E.M, Anil, B, Riedinger, C, Endicott, J.A. | | Deposit date: | 2012-10-07 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of an MDM2-Nutlin-3a complex solved by the use of a validated MDM2 surface-entropy reduction mutant.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4PHI
 
 | | Crystal structure of HEWL with hexatungstotellurate(VI) | | Descriptor: | 6-tungstotellurate(VI), ACETATE ION, GLYCEROL, ... | | Authors: | Bijelic, A, Molitor, C, Mauracher, S.G, Al-Oweini, R, Kortz, U, Rompel, A. | | Deposit date: | 2014-05-06 | | Release date: | 2015-01-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | Hen Egg-White Lysozyme Crystallisation: Protein Stacking and Structure Stability Enhanced by a Tellurium(VI)-Centred Polyoxotungstate.
Chembiochem, 16, 2015
|
|
1R51
 
 | | URATE OXIDASE FROM ASPERGILLUS FLAVUS COMPLEXED WITH ITS INHIBITOR 8-AZAXANTHIN | | Descriptor: | 8-AZAXANTHINE, CYSTEINE, Uricase | | Authors: | Prange, T, Retailleau, P, Colloc'h, N. | | Deposit date: | 2003-10-09 | | Release date: | 2004-03-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Complexed and ligand-free high-resolution structures of urate oxidase (Uox) from Aspergillus flavus: a reassignment of the active-site binding mode.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5OQ1
 
 | | Crystal structure of Serratia marcescens ChiX (used as MR model for superior PDB 5OPZ) | | Descriptor: | CHLORIDE ION, ChiX, ZINC ION | | Authors: | Owen, R.A, Fyfe, P.K, Lodge, A, Biboy, J, Vollmer, W, Hunter, W.N, Sargent, F. | | Deposit date: | 2017-08-10 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure and activity of ChiX: a peptidoglycan hydrolase required for chitinase secretion by Serratia marcescens.
Biochem. J., 475, 2018
|
|
4HFZ
 
 | | Crystal Structure of an MDM2/P53 Peptide Complex | | Descriptor: | Cellular tumor antigen p53, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Anil, B, Riedinger, C, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | The structure of an MDM2-Nutlin-3a complex solved by the use of a validated MDM2 surface-entropy reduction mutant.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3FHO
 
 | | Structure of S. pombe Dbp5 | | Descriptor: | ATP-dependent RNA helicase dbp5 | | Authors: | Cheng, Z, Song, H. | | Deposit date: | 2008-12-09 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Solution and crystal structures of mRNA exporter Dbp5p and its interaction with nucleotides
J.Mol.Biol., 388, 2009
|
|
2KQO
 
 | |
4CRP
 
 | | Solution structure of a TrkAIg2 domain construct for use in drug discovery | | Descriptor: | HIGH AFFINITY NERVE GROWTH FACTOR RECEPTOR | | Authors: | Shoemark, D.K, Fahey, M, Williams, C, Sessions, R.B, Crump, M.P, Allen-Birt, S.J. | | Deposit date: | 2014-02-28 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Design and Nuclear Magnetic Resonance (NMR) Structure Determination of the Second Extracellular Immunoglobulin Tyrosine Kinase a (Trkaig2) Domain Construct for Binding Site Elucidation in Drug Discovery
J.Med.Chem., 58, 2015
|
|
4IDW
 
 | | Polycrystalline T6 Bovine Insulin: Anisotropic Lattice Evolution and Novel Structure Refinement Strategy | | Descriptor: | Insulin A chain, Insulin B chain, ZINC ION | | Authors: | Margiolaki, I, Giannopoulou, A.E, Wright, J.P, Knight, L, Norrman, M, Schluckebier, G, Fitch, A, Von Dreele, R.B. | | Deposit date: | 2012-12-13 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-30 | | Method: | POWDER DIFFRACTION | | Cite: | High-resolution powder X-ray data reveal the T6 hexameric form of bovine insulin
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2KXW
 
 | |
2KPY
 
 | | Solution Structure of the major allergen of Artemisia vulgaris (Art v 1) | | Descriptor: | Major pollen allergen Art v 1 | | Authors: | Razzera, G, Gadermaier, G, Almeida, M, Egger, M, Jahn-Schmid, B, Almeida, F, Ferreira, F, Valente, A. | | Deposit date: | 2009-10-23 | | Release date: | 2010-10-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Mapping the interactions between a major pollen allergen and human IgE antibodies.
Structure, 18, 2010
|
|
5V2C
 
 | | RE-REFINEMENT OF CRYSTAL STRUCTURE OF PHOTOSYSTEM II COMPLEX | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Wang, J, Wiwczar, J.M, Brudvig, G.W. | | Deposit date: | 2017-03-03 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chlorophyll a with a farnesyl tail in thermophilic cyanobacteria.
Photosyn. Res., 134, 2017
|
|
1JT9
 
 | | Structure of the mutant F174A T form of the Glucosamine-6-Phosphate deaminase from E.coli | | Descriptor: | Glucosamine-6-Phosphate deaminase | | Authors: | Bustos-Jaimes, I, Sosa-Peinado, A, Rudino-Pinera, E, Horjales, E, Calcagno, M.L. | | Deposit date: | 2001-08-20 | | Release date: | 2002-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | On the role of the conformational flexibility of the active-site lid on the allosteric kinetics of glucosamine-6-phosphate deaminase.
J.Mol.Biol., 319, 2002
|
|
4DG7
 
 | |
1J2O
 
 | | Structure of FLIN2, a complex containing the N-terminal LIM domain of LMO2 and ldb1-LID | | Descriptor: | Fusion of Rhombotin-2 and LIM domain-binding protein 1, ZINC ION | | Authors: | Deane, J.E, Mackay, J.P, Kwan, A.H, Sum, E.Y, Visvader, J.E, Matthews, J.M. | | Deposit date: | 2003-01-08 | | Release date: | 2003-05-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of ldb1 by the N-terminal LIM domains of LMO2 and LMO4
EMBO J., 22, 2003
|
|
1K4U
 
 | | Solution structure of the C-terminal SH3 domain of p67phox complexed with the C-terminal tail region of p47phox | | Descriptor: | PHAGOCYTE NADPH OXIDASE SUBUNIT P47PHOX, PHAGOCYTE NADPH OXIDASE SUBUNIT P67PHOX | | Authors: | Kami, K, Takeya, R, Sumimoto, H, Kohda, D. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Diverse recognition of non-PxxP peptide ligands by the SH3 domains from p67(phox), Grb2 and Pex13p.
EMBO J., 21, 2002
|
|
4BIM
 
 | |
4OM9
 
 | | X-Ray Crystal Structure of the passenger domain of Plasmid encoded toxin, an Autrotansporter Enterotoxin from enteroaggregative Escherichia coli (EAEC) | | Descriptor: | Serine protease pet | | Authors: | Meza-Aguilar, J.D, Fromme, P, Torres-Larios, A, Mendoza-Hernandez, G, Hernandez-Chinas, U, Arreguin-Espinosa de Los Monteros, R.A, Eslava-Campos, C.A, Fromme, R. | | Deposit date: | 2014-01-27 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of the passenger domain of plasmid encoded toxin(Pet), an autotransporter enterotoxin from enteroaggregative Escherichia coli (EAEC).
Biochem.Biophys.Res.Commun., 445, 2014
|
|
