5HGM
 
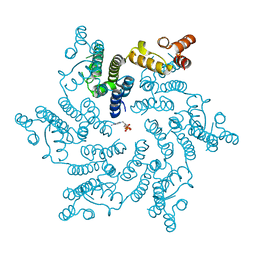 | | Hexameric HIV-1 CA in complex with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Capsid protein P24 | | Authors: | Jacques, D.A, James, L.C. | | Deposit date: | 2016-01-08 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis.
Nature, 536, 2016
|
|
8FGM
 
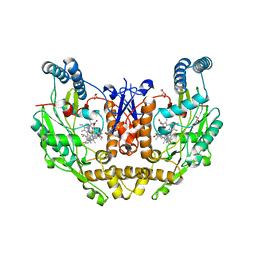 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 4-(difluoromethyl)-6-(5-(2-(dimethylamino)ethyl)-2,3-difluorophenethyl)pyridin-2-amine | | Descriptor: | 4-(difluoromethyl)-6-(2-{5-[2-(dimethylamino)ethyl]-2,3-difluorophenyl}ethyl)pyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-12-12 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Potent, Selective, and Membrane Permeable 2-Amino-4-Substituted Pyridine-Based Neuronal Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 66, 2023
|
|
6W0E
 
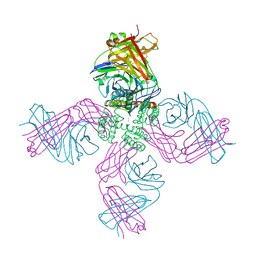 | | Open-gate KcsA soaked in 10 mM BaCl2 | | Descriptor: | BARIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.512 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
8FGA
 
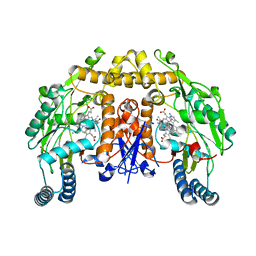 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(5-(2-(dimethylamino)ethyl)-2,3-difluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{5-[2-(dimethylamino)ethyl]-2,3-difluorophenyl}ethyl)-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-12-12 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8947 Å) | | Cite: | Potent, Selective, and Membrane Permeable 2-Amino-4-Substituted Pyridine-Based Neuronal Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 66, 2023
|
|
8ZER
 
 | | Crystal structure of the complex of Wuhan SARS-CoV-2 RBD (319-541) with P2C5 nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody P2C5, Spike protein S1, ... | | Authors: | Sluchanko, N.N, Varfolomeeva, L.A, Shcheblyakov, D.V, Logunov, D.Y, Gintsburg, A.L, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-05-06 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Evasion of New SARS-CoV-2 Variants from the Potent Virus-Neutralizing Nanobody Targeting the S-Protein Receptor-Binding Domain.
Biochemistry Mosc., 89, 2024
|
|
4ZHE
 
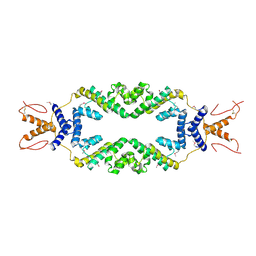 | | Crystal structure of the SeMet substituted Topless related protein 2 (TPR2) N-terminal domain (1-209) from rice | | Descriptor: | ASPR2 protein | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-05 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors.
Sci Adv, 1, 2015
|
|
8FTT
 
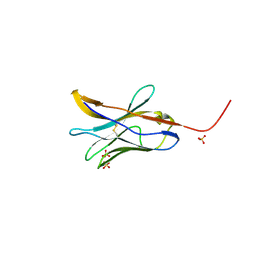 | |
8FGK
 
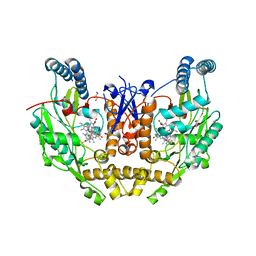 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 6-(5-(2-(diethylamino)ethyl)-2,3-difluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{5-[2-(diethylamino)ethyl]-2,3-difluorophenyl}ethyl)-4-methylpyridin-2-amine, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-12-12 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Potent, Selective, and Membrane Permeable 2-Amino-4-Substituted Pyridine-Based Neuronal Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 66, 2023
|
|
4ZRD
 
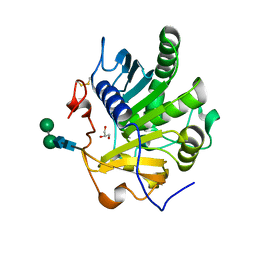 | | Crystal structure of SMG1 F278N mutant | | Descriptor: | GLYCEROL, LIP1, secretory lipase (Family 3), ... | | Authors: | Xu, J, Xu, H, Hou, S, Liu, J. | | Deposit date: | 2015-05-12 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of product-bound SMG1 lipase: active site gating implications.
Febs J., 282, 2015
|
|
8FGU
 
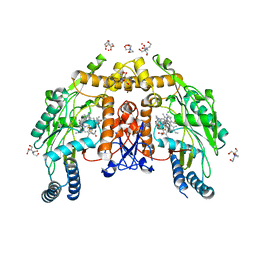 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 4-(difluoromethyl)-6-(5-(2-(dimethylamino)ethyl)-2,3-difluorophenethyl)pyridin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(difluoromethyl)-6-(2-{5-[2-(dimethylamino)ethyl]-2,3-difluorophenyl}ethyl)pyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-12-12 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent, Selective, and Membrane Permeable 2-Amino-4-Substituted Pyridine-Based Neuronal Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 66, 2023
|
|
8HUK
 
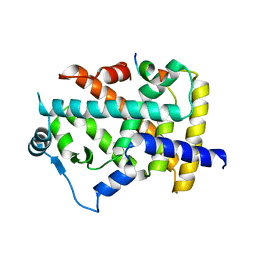 | | X-ray structure of human PPAR alpha ligand binding domain-lanifibranor-SRC1 coactivator peptide co-crystals obtained by soaking | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 4-[1-(1,3-benzothiazol-6-ylsulfonyl)-5-chloro-indol-2-yl]butanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Honda, A, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.981 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8FAJ
 
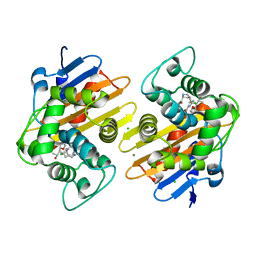 | | OXA-48-NA-1-157 inhibitor complex | | Descriptor: | (5R)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, CADMIUM ION, ... | | Authors: | Smith, C.A, Stewart, N.K, Toth, M, Vakulenko, S.B. | | Deposit date: | 2022-11-28 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The C5 alpha-Methyl-Substituted Carbapenem NA-1-157 Exhibits Potent Activity against Klebsiella spp. Isolates Producing OXA-48-Type Carbapenemases.
Acs Infect Dis., 9, 2023
|
|
8HUQ
 
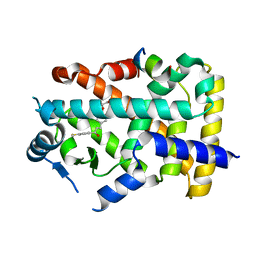 | | X-ray structure of human PPAR alpha ligand binding domain-elafibranor-SRC1 coactivator peptide co-crystals obtained by soaking | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[2,6-dimethyl-4-[(~{E})-3-(4-methylsulfanylphenyl)-3-oxidanylidene-prop-1-enyl]phenoxy]-2-methyl-propanoic acid, GLYCEROL, ... | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Honda, A, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
5HAI
 
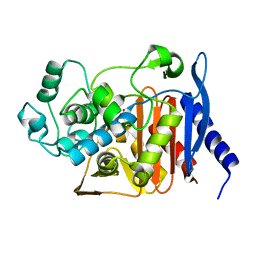 | | P99 beta-lactamase mutant - S64G | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Stojanoski, V, Adamski, C.J, Hu, L, Mehta, S.C, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2015-12-30 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Removal of the Side Chain at the Active-Site Serine by a Glycine Substitution Increases the Stability of a Wide Range of Serine beta-Lactamases by Relieving Steric Strain.
Biochemistry, 55, 2016
|
|
8FGD
 
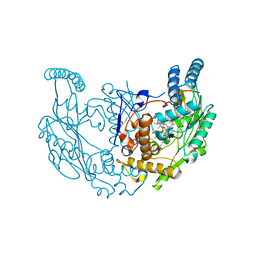 | | Structure of rat neuronal nitric oxide synthase R349A mutant heme domain in complex with 6-(5-(2-(diethylamino)ethyl)-2,3-difluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{5-[2-(diethylamino)ethyl]-2,3-difluorophenyl}ethyl)-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-12-12 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Potent, Selective, and Membrane Permeable 2-Amino-4-Substituted Pyridine-Based Neuronal Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 66, 2023
|
|
5IFN
 
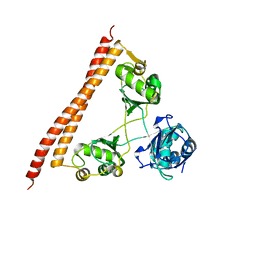 | |
8HUL
 
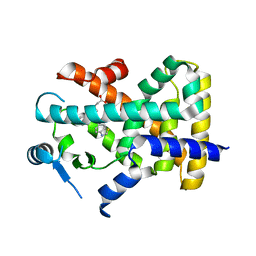 | | X-ray structure of human PPAR delta ligand binding domain-lanifibranor co-crystals obtained by co-crystallization | | Descriptor: | 4-[1-(1,3-benzothiazol-6-ylsulfonyl)-5-chloro-indol-2-yl]butanoic acid, Peroxisome proliferator-activated receptor delta | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8FGP
 
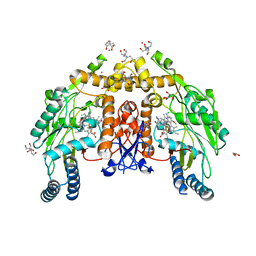 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 6-(5-(2-(dimethylamino)ethyl)-2,3-difluorophenethyl)-4-methoxypyridin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{5-[2-(dimethylamino)ethyl]-2,3-difluorophenyl}ethyl)-4-methoxypyridin-2-amine, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-12-12 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Potent, Selective, and Membrane Permeable 2-Amino-4-Substituted Pyridine-Based Neuronal Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 66, 2023
|
|
6W3C
 
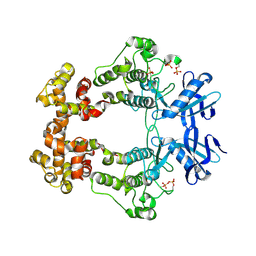 | | Structure of phosphorylated apo IRE1 | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Wallweber, H, Mortara, K, Ferri, E, Wang, W, Rudolph, J. | | Deposit date: | 2020-03-09 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Activation of the IRE1 RNase through remodeling of the kinase front pocket by ATP-competitive ligands.
Nat Commun, 11, 2020
|
|
8HUN
 
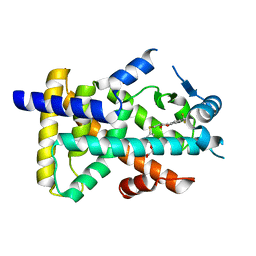 | | X-ray structure of human PPAR alpha ligand binding domain-seladelpar co-crystals obtained by cross-seeding | | Descriptor: | GLYCEROL, Peroxisome proliferator-activated receptor alpha, Seladelpar | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Iino, S, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
5IFT
 
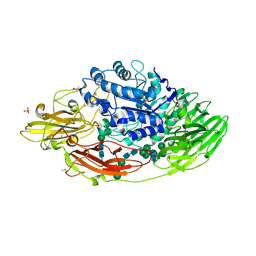 | | STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 3-b-Galactopyranosyl glucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Rico-Diaz, A, Ramirez-Escudero, M, Vizoso Vazquez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2016-02-26 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural features of Aspergillus niger beta-galactosidase define its activity against glycoside linkages.
FEBS J., 284, 2017
|
|
6W3X
 
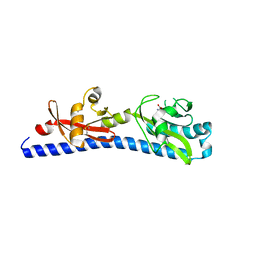 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-valine | | Descriptor: | GLYCEROL, Methyl-accepting chemotaxis protein, SULFATE ION, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
5IG1
 
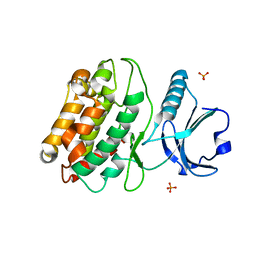 | | Crystal structure of S. rosetta CaMKII kinase domain | | Descriptor: | CAMK/CAMK2 protein kinase, PHOSPHATE ION | | Authors: | Bhattacharyya, M, Gee, C.L, Barros, T, Kuriyan, J. | | Deposit date: | 2016-02-26 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular mechanism of activation-triggered subunit exchange in Ca(2+)/calmodulin-dependent protein kinase II.
Elife, 5, 2016
|
|
8FGL
 
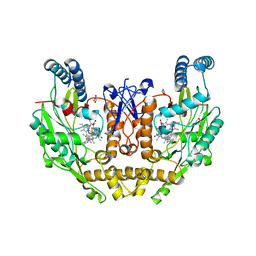 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 6-(2,3-difluoro-5-(2-(4-methylpiperazin-1-yl)ethyl)phenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{2,3-difluoro-5-[2-(4-methylpiperazin-1-yl)ethyl]phenyl}ethyl)-4-methylpyridin-2-amine, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-12-12 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Potent, Selective, and Membrane Permeable 2-Amino-4-Substituted Pyridine-Based Neuronal Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 66, 2023
|
|
8HUP
 
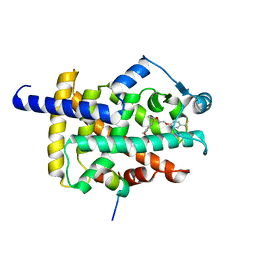 | | X-ray structure of human PPAR gamma ligand binding domain-seladelpar-SRC1 coactivator peptide co-crystals obtained by co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, Isoform 1 of Peroxisome proliferator-activated receptor gamma, Seladelpar | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
