7WJ2
 
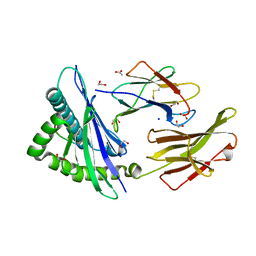 | | Crystal structure of HLA-C*1402 complexed with 8-mer HIV gag peptide | | Descriptor: | 1,2-ETHANEDIOL, 8-mer peptide, ACETATE ION, ... | | Authors: | Kuroha, J, Morita, D, Asa, M, Sugita, M. | | Deposit date: | 2022-01-05 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structures of N-myristoylated lipopeptide-bound HLA class I complexes indicate reorganization of B-pocket architecture upon ligand binding.
J.Biol.Chem., 298, 2022
|
|
7WMK
 
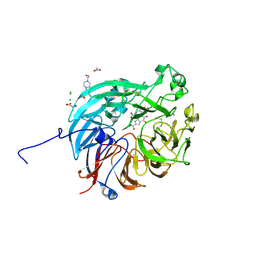 | | PQQ-dependent alcohol dehydrogenase complexed with PQQ | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, CALCIUM ION, ... | | Authors: | Chen, M, Yang, H, Lv, F. | | Deposit date: | 2022-01-15 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure-Function Analysis of a Quinone-Dependent Dehydrogenase Capable of Deoxynivalenol Detoxification.
J.Agric.Food Chem., 70, 2022
|
|
6ZJU
 
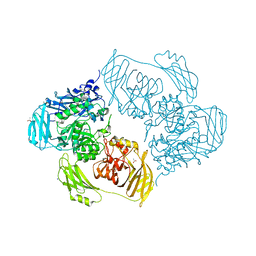 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E441Q in complex with saccharose | | Descriptor: | ACETATE ION, Beta-galactosidase, MALONATE ION, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mapping the Transglycosylation Relevant Sites of Cold-Adapted beta-d-Galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 21, 2020
|
|
6E7T
 
 | |
4MLM
 
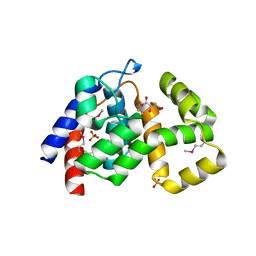 | | Crystal Structure of PhnZ from uncultured bacterium HF130_AEPn_1 | | Descriptor: | FE (III) ION, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | van Staalduinen, L.M, McSorley, F.R, Zechel, D.L, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2013-09-06 | | Release date: | 2014-04-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of PhnZ in complex with substrate reveals a di-iron oxygenase mechanism for catabolism of organophosphonates.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6EJ8
 
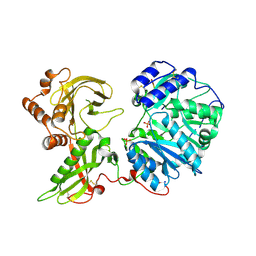 | |
7W61
 
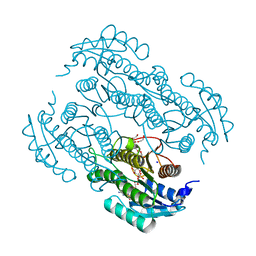 | | Crystal structure of farnesol dehydrogenase from Helicoverpa armigera | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Farnesol dehydrogenase, ... | | Authors: | Kumar, R, Das, J, Mahto, J.K, Sharma, M, Kumar, P, Sharma, A.K. | | Deposit date: | 2021-12-01 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure and molecular characterization of NADP + -farnesol dehydrogenase from cotton bollworm, Helicoverpaarmigera.
Insect Biochem.Mol.Biol., 147, 2022
|
|
4MMF
 
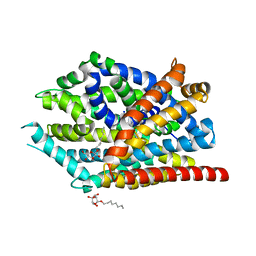 | | Crystal structure of LeuBAT (delta5 mutant) in complex with mazindol | | Descriptor: | (5R)-5-(4-chlorophenyl)-2,5-dihydro-3H-imidazo[2,1-a]isoindol-5-ol, SODIUM ION, Transporter, ... | | Authors: | Wang, H, Gouaux, E. | | Deposit date: | 2013-09-08 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for action by diverse antidepressants on biogenic amine transporters.
Nature, 503, 2013
|
|
4MS2
 
 | | Structural basis of Ca2+ selectivity of a voltage-gated calcium channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein | | Authors: | Tang, L, Gamal El-Din, T.M, Payandeh, J, Martinez, G.Q, Heard, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2013-09-18 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for Ca2+ selectivity of a voltage-gated calcium channel.
Nature, 505, 2014
|
|
4MTF
 
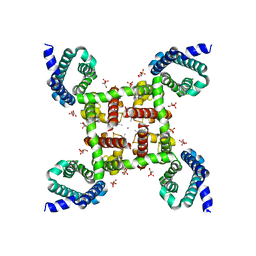 | | Structural Basis of Ca2+ Selectivity of a Voltage-gated Calcium Channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein | | Authors: | Tang, L, Gamal El-Din, T.M, Payandeh, J, Martinez, G.Q, Heard, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2013-09-19 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for Ca2+ selectivity of a voltage-gated calcium channel.
Nature, 505, 2014
|
|
6YZ2
 
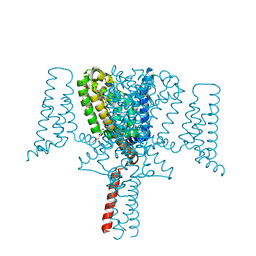 | | Full length Open-form Sodium Channel NavMs F208L | | Descriptor: | DODECAETHYLENE GLYCOL, HEGA-10, Ion transport protein, ... | | Authors: | Sula, A, Sait, L.G, Hollingworth, D, Wallace, B.A. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cannabidiol interactions with voltage-gated sodium channels.
Elife, 9, 2020
|
|
7VYY
 
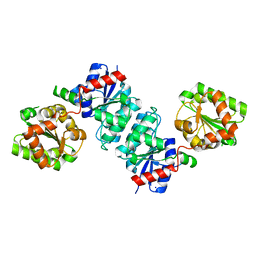 | | The crystal structure of Non-hydrolyzing UDPGlcNAc 2-epimerase | | Descriptor: | Putative UDP-N-acetylglucosamine 2-epimerase, SODIUM ION | | Authors: | Li, T.L, Rattinam, R. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.443 Å) | | Cite: | KasQ an Epimerase Primes the Biosynthesis of Aminoglycoside Antibiotic Kasugamycin and KasF/H Acetyltransferases Inactivate Its Activity.
Biomedicines, 10, 2022
|
|
7VZA
 
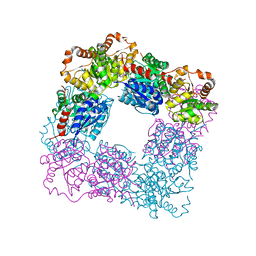 | |
7VZ6
 
 | |
6ZJQ
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E517Q in complex with galactose | | Descriptor: | ACETATE ION, Beta-galactosidase, MALONATE ION, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mapping the Transglycosylation Relevant Sites of Cold-Adapted beta-d-Galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 21, 2020
|
|
6ZJX
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant D207A in complex with saccharose | | Descriptor: | ACETATE ION, Beta-galactosidase, MALONATE ION, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Mapping the Transglycosylation Relevant Sites of Cold-Adapted beta-d-Galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 21, 2020
|
|
1TXZ
 
 | | Crystal structure of yeast ymx7, an ADP-ribose-1''-monophosphatase, complexed with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, Hypothetical 32.1 kDa protein in ADH3-RCA1 intergenic region, ... | | Authors: | Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-06 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and mechanism of ADP-ribose-1''-monophosphatase (Appr-1''-pase), a ubiquitous cellular processing enzyme.
Protein Sci., 14, 2005
|
|
6Z2E
 
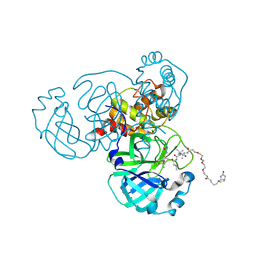 | | Crystal structure of SARS-CoV-2 Mpro in complex with the activity-based probe, biotin-PEG(4)-Abu-Tle-Leu-Gln-vinylsulfone | | Descriptor: | (4~{S})-4-[[(2~{S})-2-[[(2~{S})-2-[[(2~{S})-2-[3-[2-[2-[2-[2-[5-[(3~{a}~{S},4~{R},6~{a}~{R})-2-oxidanylidene-3,3~{a},4,6~{a}-tetrahydro-1~{H}-thieno[3,4-d]imidazol-4-yl]pentanoylamino]ethoxy]ethoxy]ethoxy]ethoxy]propanoylamino]butanoyl]amino]-3,3-dimethyl-butanoyl]amino]-4-methyl-pentanoyl]amino]-6-methylsulfonyl-hexanamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Zhang, L, Hilgenfeld, R. | | Deposit date: | 2020-05-15 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of SARS-CoV-2 Mpro in complex with the activity-based probe, biotin-PEG(4)-Abu-Tle-Leu-Gln-vinylsulfone
To Be Published
|
|
6Z31
 
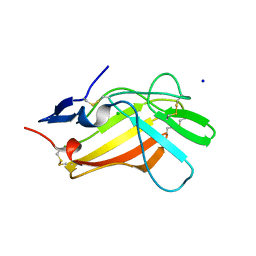 | |
4MMB
 
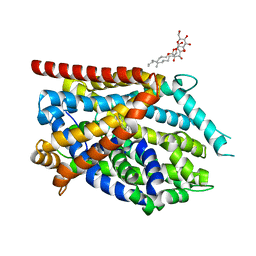 | | Crystal structure of LeuBAT (delta6 mutant) in complex with sertraline | | Descriptor: | (1S,4S)-4-(3,4-dichlorophenyl)-N-methyl-1,2,3,4-tetrahydronaphthalen-1-amine, SODIUM ION, Transporter, ... | | Authors: | Wang, H, Gouaux, E. | | Deposit date: | 2013-09-08 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for action by diverse antidepressants on biogenic amine transporters.
Nature, 503, 2013
|
|
6Z8C
 
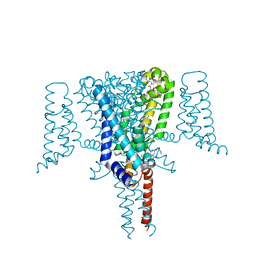 | | Crystal Structure of the Voltage-Gated Sodium Channel NavMs (F208L) in complex with N-desmethyltamoxifen (3.2 A resolution) | | Descriptor: | 2-[4-[(~{Z})-1,2-diphenylbut-1-enyl]phenoxy]-~{N}-methyl-ethanamine, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Sula, A, Hollingworth, D, Wallace, B.A. | | Deposit date: | 2020-06-02 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A tamoxifen receptor within a voltage-gated sodium channel.
Mol.Cell, 81, 2021
|
|
6E7U
 
 | |
4MTG
 
 | | Structural Basis for Ca2+ Selectivity of a Voltage-gated Calcium Channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein | | Authors: | Tang, L, Gamal El-Din, T.M, Payandeh, J, Martinez, G.Q, Heard, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2013-09-19 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.296 Å) | | Cite: | Structural basis for Ca2+ selectivity of a voltage-gated calcium channel.
Nature, 505, 2014
|
|
6E7S
 
 | |
4MVM
 
 | | Structural Basis for Ca2+ Selectivity of a Voltage-gated Calcium Channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein | | Authors: | Tang, L, Gamal El-Din, T.M, Payandeh, J, Martinez, G.Q, Heard, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2013-09-24 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1997 Å) | | Cite: | Structural basis for Ca2+ selectivity of a voltage-gated calcium channel.
Nature, 505, 2014
|
|
