7UID
 
 | | Thyclotides peptide nucleic acid in complex with DNA | | Descriptor: | CHLORIDE ION, DNA (5'-D(*GP*AP*TP*GP*TP*GP*AP*TP*A)-3'), Thyclotide, ... | | Authors: | Botos, I, Appella, D.H. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Variation of Tetrahydrofurans in Thyclotides Enhances Oligonucleotide Binding and Cellular Uptake of Peptide Nucleic Acids.
Jacs Au, 3, 2023
|
|
6JYK
 
 | | Crystal Structure of C. crescentus free GapR | | Descriptor: | UPF0335 protein CCNA_03428 | | Authors: | Xia, B, Huang, Q. | | Deposit date: | 2019-04-26 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | GapR binds DNA through dynamic opening of its tetrameric interface.
Nucleic Acids Res., 48, 2020
|
|
2KCA
 
 | | GP16 | | Descriptor: | Bacteriophage SPP1 complete nucleotide sequence | | Authors: | Gallopin, M, Gilquin, B, Zinn-Justin, S. | | Deposit date: | 2008-12-18 | | Release date: | 2009-06-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7QD5
 
 | | Cryo-EM structure of Tn4430 TnpA transposase from Tn3 family in complex with 48 bp long transposon end DNA | | Descriptor: | IR48 DNA substrate, non transferred strand, IR48 transferred strand, ... | | Authors: | Shkumatov, A.V, Oger, C.A, Aryanpour, N, Hallet, B.F, Efremov, R.G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into Tn3 family transposition mechanism.
Nat Commun, 13, 2022
|
|
2KBZ
 
 | |
1RRR
 
 | | RNA DUPLEX CONTAINING A PURINE-RICH STRAND, NMR, 10 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*AP*AP*GP*AP*GP*AP*AP*GP*C)-3'), RNA (5'-R(*GP*CP*UP*UP*CP*UP*CP*UP*UP*C)-3') | | Authors: | Gyi, J.I, Lane, A.N, Conn, G.L, Brown, T. | | Deposit date: | 1997-10-21 | | Release date: | 1998-04-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of DNA.RNA hybrids with purine-rich and pyrimidine-rich strands: comparison with the homologous DNA and RNA duplexes.
Biochemistry, 37, 1998
|
|
1RZ9
 
 | | Crystal Structure of AAV Rep complexed with the Rep-binding sequence | | Descriptor: | 26-MER, Rep protein | | Authors: | Hickman, A.B, Ronning, D.R, Perez, Z.N, Kotin, R.M, Dyda, F. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The nuclease domain of adeno-associated virus rep coordinates replication initiation using two distinct DNA recognition interfaces.
Mol.Cell, 13, 2004
|
|
454D
 
 | | INTERCALATION AND MAJOR GROOVE RECOGNITION IN THE 1.2 A RESOLUTION CRYSTAL STRUCTURE OF RH[ME2TRIEN]PHI BOUND TO 5'-G(5IU)TGCAAC-3' | | Descriptor: | 5'-D(*GP*(5IU)P*TP*GP*CP*AP*AP*C)-3', DELTA-ALPHA-RH[2R,9R-DIAMINO-4,7-DIAZADECANE]9,10-PHENANTHRENEQUINONE DIIMINE | | Authors: | Kielkopf, C.L, Erkkila, K.E, Hudson, B.P, Barton, J.K, Rees, D.C. | | Deposit date: | 1999-03-03 | | Release date: | 2000-02-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of a photoactive rhodium complex intercalated into DNA.
Nat.Struct.Biol., 7, 2000
|
|
2P6U
 
 | |
8BT1
 
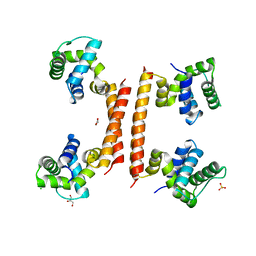 | | YdaT transcription regulator (CII functional analog) | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Prolic-Kalinsek, M, Loris, R. | | Deposit date: | 2022-11-27 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.39788437 Å) | | Cite: | Structural basis of DNA binding by YdaT, a functional equivalent of the CII repressor in the cryptic prophage CP-933P from Escherichia coli O157:H7.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
1GXC
 
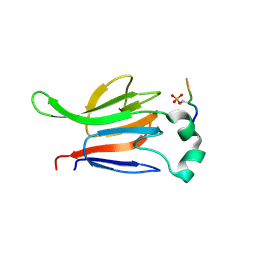 | | FHA domain from human Chk2 kinase in complex with a synthetic phosphopeptide | | Descriptor: | SERINE/THREONINE-PROTEIN KINASE CHK2, SYNTHETIC PHOSPHOPEPTIDE | | Authors: | Li, J, Williams, B.L, Haire, L.F, Goldberg, M, Wilker, E, Durocher, D, Yaffe, M.B, Jackson, S.P, Smerdon, S.J. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-13 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Versatility of the Fha Domain in DNA-Damage Signaling by the Tumor Suppressor Kinase Chk2
Mol.Cell, 9, 2002
|
|
3HKM
 
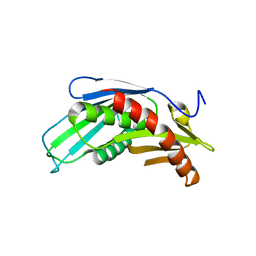 | | Crystal Structure of rice(Oryza sativa) Rrp46 | | Descriptor: | Os03g0854200 protein | | Authors: | Yang, C.-C, Wang, Y.-T, Hsiao, Y.-Y, Doudeva, L.G, Yuan, H.S. | | Deposit date: | 2009-05-25 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9845 Å) | | Cite: | Structural and biochemical characterization of CRN-5 and Rrp46: an exosome component participating in apoptotic DNA degradation
Rna, 16, 2010
|
|
5DAZ
 
 | |
2GYX
 
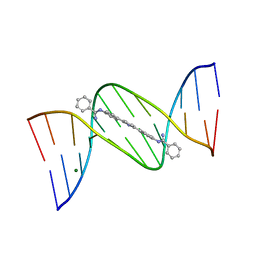 | | Crystal structure of DB884- D(CGCGAATTCGCG)2 complex at 1.86 A resolution. | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION, N,N'-(1H-PYRROLE-2,5-DIYLDI-4,1-PHENYLENE)DIBENZENECARBOXIMIDAMIDE | | Authors: | Lee, M.P.H, Neidle, S. | | Deposit date: | 2006-05-10 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Induced fit conformational changes of a "reversed amidine" heterocycle: optimized interactions in a DNA minor groove complex.
J.Am.Chem.Soc., 129, 2007
|
|
1T05
 
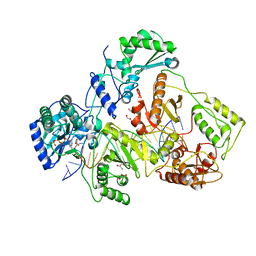 | | HIV-1 reverse transcriptase crosslinked to template-primer with tenofovir-diphosphate bound as the incoming nucleotide substrate | | Descriptor: | GLYCEROL, MAGNESIUM ION, POL polyprotein, ... | | Authors: | Tuske, S, Sarafianos, S.G, Ding, J, Arnold, E. | | Deposit date: | 2004-04-07 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of HIV-1 RT-DNA complexes before and after incorporation of the anti-AIDS drug tenofovir
Nat.Struct.Mol.Biol., 11, 2004
|
|
3KTZ
 
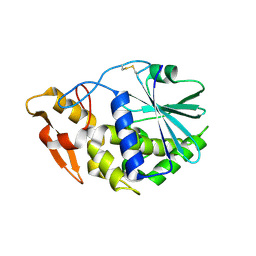 | | Structure of GAP31 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome-inactivating protein gelonin | | Authors: | Kong, X.-P. | | Deposit date: | 2009-11-26 | | Release date: | 2010-01-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new activity of anti-HIV and anti-tumor protein GAP31: DNA adenosine glycosidase--structural and modeling insight into its functions.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
2K97
 
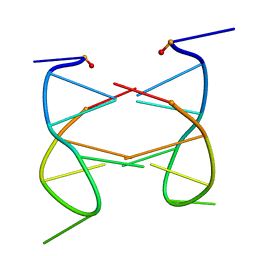 | | Dimeric solution structure of the cyclic octamer d(pCGCTCCGT) | | Descriptor: | 5'-D(P*CP*GP*CP*TP*CP*CP*GP*T)-3' | | Authors: | Viladoms, J, Escaja, N, Frieden, M, Gomez-Pinto, I, Pedroso, E, Gonzalez, C. | | Deposit date: | 2008-09-30 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Self-association of short DNA loops through minor groove C:G:G:C tetrads.
Nucleic Acids Res., 37, 2009
|
|
2ET0
 
 | | The structure of a three-way DNA junction in complex with a metallo-supramolecular helicate reveals a new target for drugs | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', FE (II) ION, N-[(1E)-PYRIDIN-2-YLMETHYLENE]-N-[4-(4-{[(1E)-PYRIDIN-2-YLMETHYLENE]AMINO}BENZYL)PHENYL]AMINE | | Authors: | Oleksi, A, Blanco, A.G, Boer, R, Uson, I, Aymami, J, Coll, M. | | Deposit date: | 2005-10-27 | | Release date: | 2006-03-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Recognition of a Three-Way DNA Junction by a Metallosupramolecular Helicate
ANGEW.CHEM.INT.ED.ENGL., 45, 2006
|
|
1R05
 
 | | Solution Structure of Max B-HLH-LZ | | Descriptor: | Max protein | | Authors: | Sauv, S, Tremblay, L, Lavigne, P. | | Deposit date: | 2003-09-19 | | Release date: | 2003-10-21 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of a mutant of the Max b/HLH/LZ free of DNA: insights into the specific and reversible DNA binding mechanism of dimeric transcription factors
J.Mol.Biol., 342, 2004
|
|
3GOM
 
 | | Barium bound to the Holliday junction sequence d(TCGGCGCCGA)4 | | Descriptor: | 5'-D(*TP*CP*GP*GP*CP*GP*CP*CP*GP*A)-3', BARIUM ION | | Authors: | Naseer, A, Cardin, C.J. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure determination of an intercalating ruthenium dipyridophenazine complex which kinks DNA by semiintercalation of a tetraazaphenanthrene ligand.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1SP2
 
 | |
1SP1
 
 | |
3GPL
 
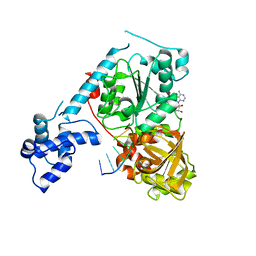 | | Crystal structure of the ternary complex of RecD2 with DNA and ADPNP | | Descriptor: | 5'-D(*T*TP*TP*TP*TP*TP*TP*T)-3', Exodeoxyribonuclease V, subunit RecD, ... | | Authors: | Saikrishnan, K, Cook, N, Wigley, D.B. | | Deposit date: | 2009-03-23 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic basis of 5'-3' translocation in SF1B helicases.
Cell(Cambridge,Mass.), 137, 2009
|
|
1SXQ
 
 | |
3V9W
 
 | |
