4Q0L
 
 | | Crystal structure of catalytic domain of human carbonic anhydrase isozyme XII with inhibitor | | Descriptor: | 3-(cyclooctylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 12, ZINC ION | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-04-02 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and characterization of novel selective inhibitors of carbonic anhydrase IX.
J.Med.Chem., 57, 2014
|
|
4Q0Y
 
 | |
4Q9J
 
 | | P-glycoprotein cocrystallised with QZ-Val | | Descriptor: | Multidrug resistance protein 1A, N-{[5-AMINO-1-(5-O-PHOSPHONO-BETA-D-ARABINOFURANOSYL)-1H-IMIDAZOL-4-YL]CARBONYL}-L-ASPARTIC ACID | | Authors: | McGrath, A.P, Szewczyk, P, Chang, G. | | Deposit date: | 2014-05-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4QHZ
 
 | |
4QK4
 
 | | Crystal structure of human nuclear receptor sf-1 (nr5a1) bound to pip2 at 2.8 a resolution | | Descriptor: | (2S)-3-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dihexadecanoate, 1,2-ETHANEDIOL, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, ... | | Authors: | Joint Center for Structural Genomics (JCSG), Partnership for Stem Cell Biology (STEMCELL) | | Deposit date: | 2014-06-05 | | Release date: | 2014-07-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The signaling phospholipid PIP3 creates a new interaction surface on the nuclear receptor SF-1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QP4
 
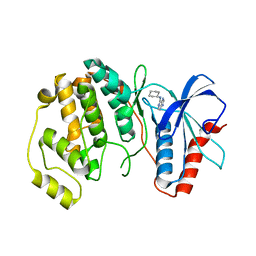 | |
4QPA
 
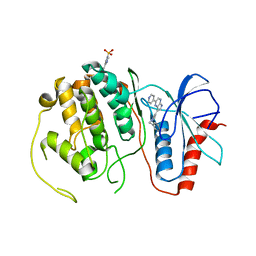 | |
4QP7
 
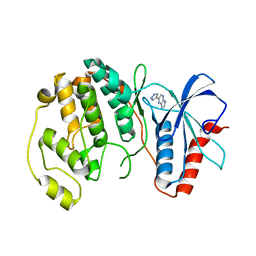 | |
4Q1Z
 
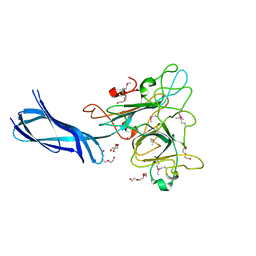 | |
4QHW
 
 | |
4QP3
 
 | |
4PQX
 
 | |
4PUX
 
 | |
4PNZ
 
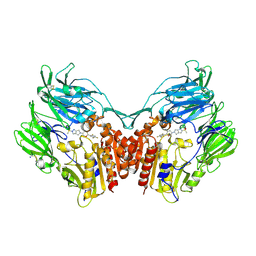 | | Human dipeptidyl peptidase IV/CD26 in complex with the long-acting inhibitor Omarigliptin (MK-3102) | | Descriptor: | (2R,3S,5R)-5-[2-(methylsulfonyl)-2,6-dihydropyrrolo[3,4-c]pyrazol-5(4H)-yl]-2-(2,4,5-trifluorophenyl)tetrahydro-2H-pyran-3-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G, Yan, Y. | | Deposit date: | 2014-02-22 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Omarigliptin (MK-3102): A Novel Long-Acting DPP-4 Inhibitor for Once-Weekly Treatment of Type 2 Diabetes.
J.Med.Chem., 57, 2014
|
|
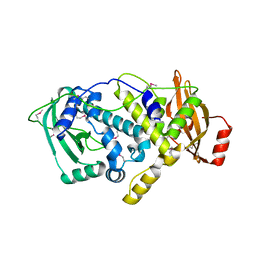
4QAN
 
 | |
4QHX
 
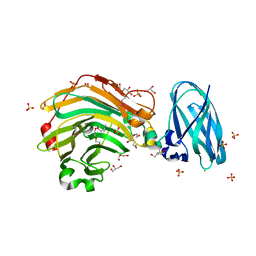 | |
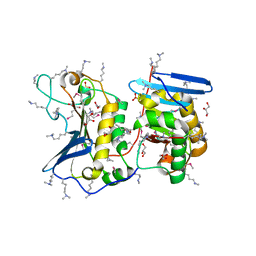
4QE0
 
 | |
4QGP
 
 | |
4QHB
 
 | |
4QIY
 
 | | Crystal structure of human carbonic anhydrase isozyme II with inhibitor | | Descriptor: | 2,3,6-trifluoro-5-{[(1R,2S)-2-hydroxy-1,2-diphenylethyl]amino}-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Manakova, E, Smirnov, A, Grazulis, S. | | Deposit date: | 2014-06-03 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Functionalization of Fluorinated Benzenesulfonamides and Their Inhibitory Properties toward Carbonic Anhydrases
Chemmedchem, 10, 2015
|
|
4QJ0
 
 | | Crystal structure of catalytic domain of human carbonic anhydrase isozyme XII with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2,3,6-trifluoro-5-{[(1R,2S)-2-hydroxy-1,2-diphenylethyl]amino}-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 12, ... | | Authors: | Manakova, E, Smirnov, A, Grazulis, S. | | Deposit date: | 2014-06-03 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Functionalization of Fluorinated Benzenesulfonamides and Their Inhibitory Properties toward Carbonic Anhydrases
Chemmedchem, 10, 2015
|
|
4QJR
 
 | | Crystal structure of human nuclear receptor sf-1 (nr5a1) bound to its hormone pip3 at 2.4 a resolution | | Descriptor: | (2S)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane -1,2-diyl dihexadecanoate, ACETATE ION, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, ... | | Authors: | Joint Center for Structural Genomics (JCSG), Partnership for Stem Cell Biology (STEMCELL) | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The signaling phospholipid PIP3 creates a new interaction surface on the nuclear receptor SF-1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QJW
 
 | | Crystal structure of catalytic domain of human carbonic anhydrase isozyme XII with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-(benzylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-06-05 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Functionalization of Fluorinated Benzenesulfonamides and Their Inhibitory Properties toward Carbonic Anhydrases
Chemmedchem, 10, 2015
|
|
4QKA
 
 | | c-di-AMP riboswitch from Thermoanaerobacter pseudethanolicus, iridium hexamine soak | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, C-di-AMP riboswitch, IRIDIUM HEXAMMINE ION, ... | | Authors: | Gao, A, Serganov, A. | | Deposit date: | 2014-06-05 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into recognition of c-di-AMP by the ydaO riboswitch.
Nat.Chem.Biol., 10, 2014
|
|
4QP6
 
 | |
