1OAK
 
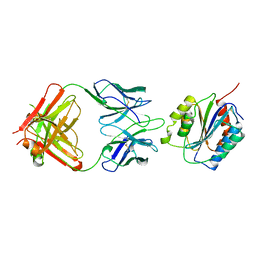 | |
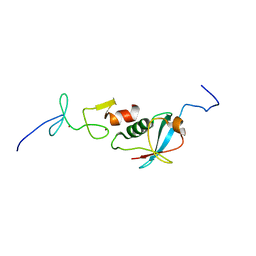
6U4N
 
 | |
3JS6
 
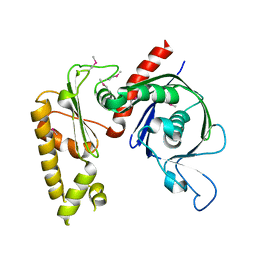 | | Crystal structure of apo psk41 parM protein | | Descriptor: | Uncharacterized ParM protein | | Authors: | Schumacher, M.A, Xu, W, Firth, N. | | Deposit date: | 2009-09-09 | | Release date: | 2010-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and filament dynamics of the pSK41 actin-like ParM protein: implications for plasmid DNA segregation.
J.Biol.Chem., 285, 2010
|
|
5WF1
 
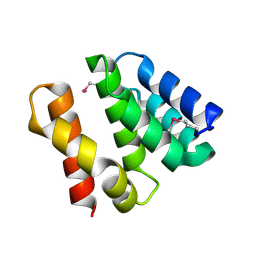 | |
5WF9
 
 | | Tepsin tENTH domain 1-153 | | Descriptor: | AP-4 complex accessory subunit Tepsin | | Authors: | Archuleta, T.L, Jackson, L.P. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and evolution of ENTH and VHS/ENTH-like domains in tepsin.
Traffic, 18, 2017
|
|
5WFB
 
 | | Tepsin tENTH domain 1-136. | | Descriptor: | AP-4 complex accessory subunit Tepsin | | Authors: | Archuleta, T.L, Jackson, L.P. | | Deposit date: | 2017-07-11 | | Release date: | 2017-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.382 Å) | | Cite: | Structure and evolution of ENTH and VHS/ENTH-like domains in tepsin.
Traffic, 18, 2017
|
|
6U7N
 
 | | Crystal structure of neurotrimin (NTM) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Neurotrimin, ... | | Authors: | Machius, M, Venkannagari, H, Misra, A, Rudenko, G, Rush, S. | | Deposit date: | 2019-09-03 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.321 Å) | | Cite: | Highly Conserved Molecular Features in IgLONs Contrast Their Distinct Structural and Biological Outcomes.
J.Mol.Biol., 432, 2020
|
|
5YCQ
 
 | | Unique Specificity-Enhancing Factor for the AAA+ Lon Protease | | Descriptor: | Heat shock protein HspQ | | Authors: | Abe, Y, Shioi, S, Kita, S, Nakata, H, Maenaka, K, Kohda, D, Katayama, T, Ueda, T. | | Deposit date: | 2017-09-08 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | X-ray crystal structure of Escherichia coli HspQ, a protein involved in the retardation of replication initiation
FEBS Lett., 591, 2017
|
|
8DVR
 
 | | Cryo-EM structure of RIG-I bound to the end of p3SLR30 (+AMPPNP) | | Descriptor: | Antiviral innate immune response receptor RIG-I, GUANOSINE-5'-TRIPHOSPHATE, ZINC ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-07-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
5NG2
 
 | | Structure of RIP2K(D146N) with bound Staurosporine | | Descriptor: | PHOSPHATE ION, Receptor-interacting serine/threonine-protein kinase 2, STAUROSPORINE | | Authors: | Pellegrini, E, Cusack, S. | | Deposit date: | 2017-03-16 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the inactive and active states of RIP2 kinase inform on the mechanism of activation.
PLoS ONE, 12, 2017
|
|
5K7L
 
 | |
8DVS
 
 | | Cryo-EM structure of RIG-I bound to the end of OHSLR30 (+ATP) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Antiviral innate immune response receptor RIG-I, MAGNESIUM ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-07-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
4IFZ
 
 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein, Mn(II)-AMP product bound form | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9012 Å) | | Cite: | The TP0796 Lipoprotein of Treponema pallidum Is a Bimetal-dependent FAD Pyrophosphatase with a Potential Role in Flavin Homeostasis.
J.Biol.Chem., 288, 2013
|
|
8DVU
 
 | |
5N76
 
 | | Crystal structure of the apo-form of the CO dehydrogenase accessory protein CooT from Rhodospirillum rubrum | | Descriptor: | CooT | | Authors: | Timm, J, Brochier-Armanet, C, Perard, J, Zambelli, B, Ollagnier-de-Choudens, S, Ciurli, S, Cavazza, C. | | Deposit date: | 2017-02-19 | | Release date: | 2017-05-10 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The CO dehydrogenase accessory protein CooT is a novel nickel-binding protein.
Metallomics, 9, 2017
|
|
5YQJ
 
 | | Crystal structure of the first StARkin domain of Lam4 | | Descriptor: | Membrane-anchored lipid-binding protein LAM4 | | Authors: | Im, Y.J, Tong, J.S. | | Deposit date: | 2017-11-06 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of sterol recognition and nonvesicular transport by lipid transfer proteins anchored at membrane contact sites
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5NMC
 
 | |
4IFU
 
 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein, apo form | | Descriptor: | FAD:protein FMN transferase, MAGNESIUM ION | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8334 Å) | | Cite: | The TP0796 Lipoprotein of Treponema pallidum Is a Bimetal-dependent FAD Pyrophosphatase with a Potential Role in Flavin Homeostasis.
J.Biol.Chem., 288, 2013
|
|
5O8O
 
 | | N. crassa Tom40 model based on cryo-EM structure of the TOM core complex at 6.8 A | | Descriptor: | Mitochondrial import receptor subunit tom40 | | Authors: | Bausewein, T, Mills, D.J, Nussberger, S, Nitschke, B, Kuehlbrandt, W. | | Deposit date: | 2017-06-13 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Cryo-EM Structure of the TOM Core Complex from Neurospora crassa.
Cell, 170, 2017
|
|
1OE9
 
 | | Crystal structure of Myosin V motor with essential light chain-nucleotide-free | | Descriptor: | MYOSIN LIGHT CHAIN 1, SLOW-TWITCH MUSCLE A ISOFORM, MYOSIN VA, ... | | Authors: | Coureux, P.-D, Wells, A.L, Menetrey, J, Yengo, C.M, Morris, C.A, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2003-03-21 | | Release date: | 2003-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Structural State of the Myosin V Motor without Bound Nucleotide
Nature, 425, 2003
|
|
6UZB
 
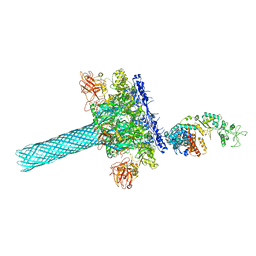 | | Anthrax toxin protective antigen channels bound to edema factor | | Descriptor: | CALCIUM ION, Calmodulin-sensitive adenylate cyclase, Protective antigen | | Authors: | Hardenbrook, N.J, Liu, S, Zhou, K, Zhou, Z.H, Krantz, B.A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Atomic structures of anthrax toxin protective antigen channels bound to partially unfolded lethal and edema factors.
Nat Commun, 11, 2020
|
|
3K4O
 
 | |
3P2Q
 
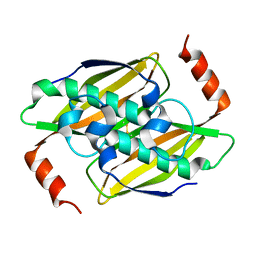 | | Crystal structure of the fluoroacetyl-CoA-specific thioesterase, FlK | | Descriptor: | Fluoroacetyl coenzyme A thioesterase | | Authors: | Weeks, A.M, Coyle, S.M, Jinek, M, Doudna, J.A, Chang, M.C.Y. | | Deposit date: | 2010-10-03 | | Release date: | 2010-10-20 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity.
Biochemistry, 49, 2010
|
|
6SEH
 
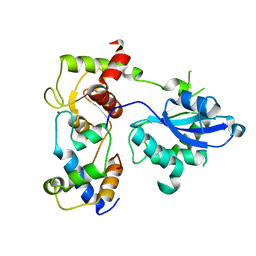 | | Recognition and processing of branched DNA substrates by Slx1-Slx4 nuclease | | Descriptor: | Structure-specific endonuclease subunit SLX1, Structure-specific endonuclease subunit SLX4, ZINC ION | | Authors: | Gaur, V, Zajko, W, Nirwal, S, Szlachcic, A, Gapinska, M, Nowotny, M. | | Deposit date: | 2019-07-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Recognition and processing of branched DNA substrates by Slx1-Slx4 nuclease.
Nucleic Acids Res., 47, 2019
|
|
3P3F
 
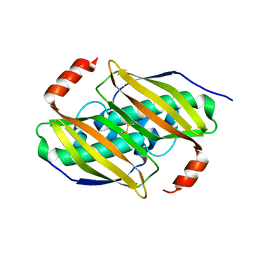 | | Crystal structure of the F36A mutant of the fluoroacetyl-CoA-specific thioesterase FlK | | Descriptor: | Fluoroacetyl coenzyme A thioesterase | | Authors: | Weeks, A.M, Coyle, S.M, Jinek, M, Doudna, J.A, Chang, M.C.Y. | | Deposit date: | 2010-10-04 | | Release date: | 2010-10-20 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity.
Biochemistry, 49, 2010
|
|
