5Z31
 
 | | LPS bound solution structure of WS2-KG18 | | Descriptor: | LYS-ASN-LYS-SER-ARG-VAL-ALA-ARG-GLY-TRP-GLY-ARG-LYS-CYS-PRO-LEU-PHE-GLY | | Authors: | Bhunia, A, Mohid, S.A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-02-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Application of tungsten disulfide quantum dot-conjugated antimicrobial peptides in bio-imaging and antimicrobial therapy.
Colloids Surf B Biointerfaces, 176, 2019
|
|
8QAQ
 
 | | Conformations of macrocyclic peptides sampled by exact NOEs: models for cell-permeability. Conformation 1 of omphalotin A in apolar solvents. | | Descriptor: | TRP-MVA-ILE-MVA-MVA-SAR-MVA-IML-SAR-VAL-IML-SAR | | Authors: | Ruedisser, S.H, Matabaro, E, Sonderegger, L, Guentert, P, Kuenzler, M, Gossert, A.D. | | Deposit date: | 2023-08-23 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | SOLUTION NMR | | Cite: | Conformations of Macrocyclic Peptides Sampled by Nuclear Magnetic Resonance: Models for Cell-Permeability.
J.Am.Chem.Soc., 145, 2023
|
|
8QAS
 
 | | Conformations of macrocyclic peptides sampled by exact NOEs: models for cell-permeability. NMR structure of Omphalotin A in methanol / water indoleOut conformation. | | Descriptor: | TRP-MVA-ILE-MVA-MVA-SAR-MVA-IML-SAR-VAL-IML-SAR | | Authors: | Ruedisser, S.H, Matabaro, E, Sonderegger, L, Guentert, P, Kuenzler, M, Gossert, A.D. | | Deposit date: | 2023-08-23 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | SOLUTION NMR | | Cite: | Conformations of Macrocyclic Peptides Sampled by Nuclear Magnetic Resonance: Models for Cell-Permeability.
J.Am.Chem.Soc., 145, 2023
|
|
8QBP
 
 | | Conformations of macrocyclic peptides sampled by exact NOEs: models for cell-permeability. NMR structure of Omphalotin A in methanol / water indoleOut conformation. | | Descriptor: | TRP-MVA-ILE-MVA-MVA-SAR-MVA-IML-SAR-VAL-IML-SAR | | Authors: | Ruedisser, S.H, Matabaro, E, Sonderegger, L, Guentert, P, Kuenzler, M, Gossert, A.D. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Conformations of Macrocyclic Peptides Sampled by Nuclear Magnetic Resonance: Models for Cell-Permeability.
J.Am.Chem.Soc., 145, 2023
|
|
6FCE
 
 | | NMR ensemble of Macrocyclic Peptidomimetic Containing Constrained a,a-dialkylated Amino Acids with Potent and Selective Activity at Human Melanocortin Receptors | | Descriptor: | ACP-HIS-DPHE-ARG-TRP-ASP-NH2 | | Authors: | Brancaccio, D, Carotenuto, A, Grieco, P, Merlino, F, Zhou, Y, Cai, M, Yousif, A.M, Di Maro, S, Novellino, E, Hruby, V.J. | | Deposit date: | 2017-12-20 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Development of Macrocyclic Peptidomimetics Containing Constrained alpha , alpha-Dialkylated Amino Acids with Potent and Selective Activity at Human Melanocortin Receptors.
J. Med. Chem., 61, 2018
|
|
8WVS
 
 | | The structure of CPPCXC motif | | Descriptor: | SER-CYS-PRO-PRO-CYS-LYS-CYS-GLY-TRP | | Authors: | Fan, S.H, Wu, C.L. | | Deposit date: | 2023-10-24 | | Release date: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Combinations of Disulfide-Directing Motifs in Tandem Enabling the De-sign and Discovery of Multicyclic Peptide Binders
To Be Published
|
|
5MXS
 
 | |
5MMK
 
 | | HYL-20 | | Descriptor: | GLY-ILE-LEU-SER-SER-LEU-TRP-LYS-LYS-LEU-LYS-LYS-ILE-ILE-ALA-LYS | | Authors: | Hexnerova, R. | | Deposit date: | 2016-12-10 | | Release date: | 2017-09-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | How proteases from Enterococcus faecalis contribute to its resistance to short alpha-helical antimicrobial peptides.
Pathog Dis, 75, 2017
|
|
5MXT
 
 | |
5MML
 
 | | HYL-20k | | Descriptor: | GLY-ILE-LEU-SER-SER-LEU-TRP-LYS-LYS-LEU-LYS-LYS-ILE-ILE-ALA-LYS | | Authors: | Hexnerova, R. | | Deposit date: | 2016-12-10 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | How proteases from Enterococcus faecalis contribute to its resistance to short alpha-helical antimicrobial peptides.
Pathog Dis, 75, 2017
|
|
6M0Y
 
 | | KR-12 analog derived from human LL-37 | | Descriptor: | LYS-ARG-ILE-VAL-LYS-ARG-ILE-LYS-LYS-TRP-LEU-ARG | | Authors: | Yun, H, Min, H.J, Lee, C.W. | | Deposit date: | 2020-02-24 | | Release date: | 2021-02-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Bactericidal Activity of KR-12 Analog Derived from Human LL-37 as a Potential Cosmetic Preservative
To Be Published
|
|
5ZYX
 
 | | Solution NMR structure of K30 peptide in 10 mM dioctanoyl phosphatidylglycerol (D8PG) | | Descriptor: | ARG-TRP-LYS-ARG-HIS-ILE-SER-GLU-GLN-LEU-ARG-ARG-ARG-ASP-ARG-LEU-GLN-ARG-GLN-ALA | | Authors: | Bhunia, A, Mohid, A, Stella, L, Calligari, P. | | Deposit date: | 2018-05-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design, Synthesis, Antibacterial Potential, and Structural Characterization of N-Acylated Derivatives of the Human Autophagy 16 Polypeptide.
Bioconjug.Chem., 30, 2019
|
|
6QXB
 
 | | NMR structure of peptide 7, characterized by a cis-4-amino-Pro residue, with a significant lower MIC on E. coli | | Descriptor: | PHE-VAL-CAP-TRP-PHE-SER-LYS-PHE-LEU-GLY-ARG-ILE-LEU-NH2 | | Authors: | Brancaccio, D, Carotenuto, A, Merlino, F, Grieco, P, Novellino, E. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Outcomes of Decorated Prolines in the Discovery of Antimicrobial Peptides from Temporin-L.
Chemmedchem, 14, 2019
|
|
6QXC
 
 | | NMR structure of peptide 8, characterized by a trans-4-cyclohexyl-Pro, with a dramatic reduction in activity on E. coli ATCC and lost effect on P. aeruginosa. | | Descriptor: | PHE-VAL-TCP-TRP-PHE-SER-LYS-PHE-LEU-GLY-ARG-ILE-LEU-NH2 | | Authors: | Brancaccio, D, Carotenuto, A, Merlino, F, Grieco, P, Novellino, E. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Outcomes of Decorated Prolines in the Discovery of Antimicrobial Peptides from Temporin-L.
Chemmedchem, 14, 2019
|
|
6OQP
 
 | | U-AITx-Ate1 | | Descriptor: | SER-LYS-TRP-ILE-CYS-ALA-ASN-ARG-SER-VAL-CYS-PRO-ILE | | Authors: | Elnahriry, K.A, Wai, D.C.C, Norton, R.S. | | Deposit date: | 2019-04-28 | | Release date: | 2019-07-31 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterisation of a novel peptide from the Australian sea anemone Actinia tenebrosa.
Toxicon, 168, 2019
|
|
6BJF
 
 | | NMR Structural and biophysical functional analysis of intracellular loop 5 of the NHE1 isoform of the Na+/H+ exchanger. | | Descriptor: | GLY-LEU-THR-TRP-PHE-ILE-ASN-LYS-PHE-ARG-ILE-VAL-LYS | | Authors: | McKay, R, Wong, K, Towle, K, Fliegel, L. | | Deposit date: | 2017-11-06 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Diverse residues of intracellular loop 5 of the Na+/H+exchanger modulate proton sensing, expression, activity and targeting.
Biochim Biophys Acta Biomembr, 1861, 2019
|
|
7TRP
 
 | | Human M4 muscarinic acetylcholine receptor complex with Gi1 and the agonist iperoxo and positive allosteric modulator LY2033298 | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-6-methoxy-4-methylthieno[2,3-b]pyridine-2-carboxamide, 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, Antibody fragment scFv16, ... | | Authors: | Vuckovic, Z, Mobbs, J.I, Belousoff, M.J, Glukhova, A, Sexton, P.M, Danev, R, Thal, D.M. | | Deposit date: | 2022-01-30 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural and dynamic mechanisms of allostery at the M4 muscarinic acetylcholine receptor
Elife, 2023
|
|
7V5E
 
 | |
8GVN
 
 | | Novel salt-resistant antimicrobial peptide, RR14 | | Descriptor: | TRP-LEU-ARG-ARG-ILE-LYS-ALA-TRP-LEU-ARG-ARG-ILE-LYS-ALA | | Authors: | Lin, T.L, Tseng, T.S, Fan, P.J. | | Deposit date: | 2022-09-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Deciphering Structure-Function Relationship Unveils Salt-Resistant Mode of Action of a Potent MRSA-Inhibiting Antimicrobial Peptide, RR14.
J.Bacteriol., 204, 2022
|
|
4TRP
 
 | | Crystal structure of monoclonal antibody against neuroblastoma associated antigen. | | Descriptor: | Heavy chain of monoclonal antibody against neuroblastoma associated antigen, Light chain of monoclonal antibody against neuroblastoma associated antigen | | Authors: | Horwacik, I, Golik, P, Grudnik, P, Zdzalik, M, Rokita, H, Dubin, G. | | Deposit date: | 2014-06-17 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis of GD2 Ganglioside and Mimetic Peptide Recognition by 14G2a Antibody.
Mol.Cell Proteomics, 14, 2015
|
|
5TRP
 
 | |
3TRP
 
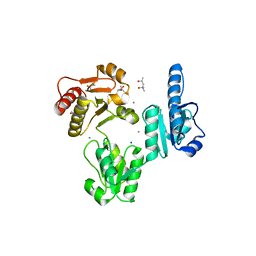 | | Crystal structure of recombinant rabbit skeletal calsequestrin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sanchez, E.J, Lewis, K.M, Munske, G.R, Nissen, M.S, Kang, C. | | Deposit date: | 2011-09-09 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8817 Å) | | Cite: | High-capacity Ca2+ Binding of Human Skeletal Calsequestrin.
J.Biol.Chem., 287, 2012
|
|
6TRP
 
 | | Solution Structure of Docking Domain Complex of Pax NRPS: PaxC NDD - PaxB CDD | | Descriptor: | Peptide synthetase XpsB,Peptide synthetase XpsB | | Authors: | Watzel, J, Hacker, C, Duchardt-Ferner, E, Bode, H.B, Woehnert, J. | | Deposit date: | 2019-12-19 | | Release date: | 2020-08-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A New Docking Domain Type in the Peptide-Antimicrobial-Xenorhabdus Peptide Producing Nonribosomal Peptide Synthetase fromXenorhabdus bovienii.
Acs Chem.Biol., 15, 2020
|
|
8TRP
 
 | |
3TRQ
 
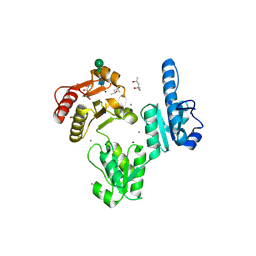 | | Crystal structure of native rabbit skeletal calsequestrin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sanchez, E.J, Lewis, K.M, Munske, G.R, Nissen, M.S, Kang, C. | | Deposit date: | 2011-09-09 | | Release date: | 2011-12-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Glycosylation of Skeletal Calsequestrin: IMPLICATIONS FOR ITS FUNCTION.
J.Biol.Chem., 287, 2012
|
|
