6GIR
 
 | | Arabidopsis thaliana cytosolic seryl-tRNA synthetase | | Descriptor: | Serine--tRNA ligase, cytoplasmic | | Authors: | Kekez, I, Kekez, M, Rokov-Plavec, J, Matkovic-Calogovic, D. | | Deposit date: | 2018-05-15 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Arabidopsis seryl-tRNA synthetase: the first crystal structure and novel protein interactor of plant aminoacyl-tRNA synthetase.
FEBS J., 286, 2019
|
|
1ICH
 
 | | SOLUTION STRUCTURE OF THE TUMOR NECROSIS FACTOR RECEPTOR-1 DEATH DOMAIN | | Descriptor: | TUMOR NECROSIS FACTOR RECEPTOR-1 | | Authors: | Sukits, S.F, Lin, L.-L, Malakian, K, Powers, R, Xu, G.-Y. | | Deposit date: | 2001-04-01 | | Release date: | 2002-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tumor necrosis factor receptor-1 death domain.
J.Mol.Biol., 310, 2001
|
|
3PY2
 
 | | Structure of C126S mutant of Plasmodium falciparum triosephosphate isomerase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Triosephosphate isomerase | | Authors: | Samanta, M, Banerjee, M, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2010-12-11 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Probing the role of the fully conserved Cys126 in triosephosphate isomerase by site-specific mutagenesis--distal effects on dimer stability.
Febs J., 278, 2011
|
|
5XLK
 
 | | Crystal structure of the flagellar cap protein FliD D2-D3 domains from Serratia marcescens in Space group I422 | | Descriptor: | Flagellar hook-associated protein 2, ZINC ION | | Authors: | Cho, S.Y, Song, W.S, Hong, H.J, Yoon, S.I. | | Deposit date: | 2017-05-10 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Tetrameric structure of the flagellar cap protein FliD from Serratia marcescens.
Biochem. Biophys. Res. Commun., 489, 2017
|
|
5X5A
 
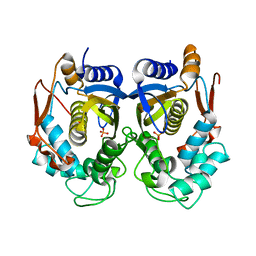 | | Human thymidylate synthase bound with phosphate ion | | Descriptor: | PHOSPHATE ION, Thymidylate synthase | | Authors: | Chen, D, Jansson, A, Larsson, A, Nordlund, P. | | Deposit date: | 2017-02-15 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural analyses of human thymidylate synthase reveal a site that may control conformational switching between active and inactive states
J. Biol. Chem., 292, 2017
|
|
4S3H
 
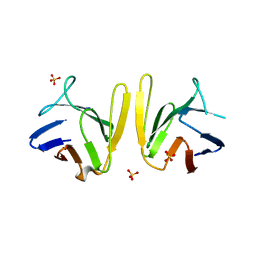 | | Crystal structure of S. pombe Mdb1 FHA domain | | Descriptor: | Mdb1, SULFATE ION | | Authors: | Luo, S, Ye, K. | | Deposit date: | 2015-01-26 | | Release date: | 2015-07-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Dimerization Mediated by a Divergent Forkhead-associated Domain Is Essential for the DNA Damage and Spindle Functions of Fission Yeast Mdb1.
J.Biol.Chem., 290, 2015
|
|
5X66
 
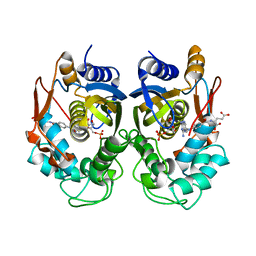 | | Human thymidylate synthase in complex with dUMP and methotrexate | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, METHOTREXATE, Thymidylate synthase | | Authors: | Chen, D, Jansson, A, Larsson, A, Nordlund, P. | | Deposit date: | 2017-02-21 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural analyses of human thymidylate synthase reveal a site that may control conformational switching between active and inactive states
J. Biol. Chem., 292, 2017
|
|
3LTM
 
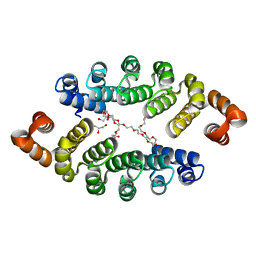 | | Structure of a new family of artificial alpha helicoidal repeat proteins (alpha-Rep) based on thermostable HEAT-like repeats | | Descriptor: | Alpha-Rep4, DODECAETHYLENE GLYCOL, GLYCEROL, ... | | Authors: | Urvoas, A, Guellouz, A, Graille, M, van Tilbeurgh, H, Desmadril, M, Minard, P. | | Deposit date: | 2010-02-16 | | Release date: | 2010-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design, production and molecular structure of a new family of artificial alpha-helicoidal repeat proteins ( alpha Rep) based on thermostable HEAT-like repeats
J.Mol.Biol., 404, 2010
|
|
3L5W
 
 | | Crystal structure of the complex between IL-13 and C836 FAB | | Descriptor: | C836 HEAVY CHAIN, C836 LIGHT CHAIN, GLYCEROL, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-22 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
3PWA
 
 | | Structure of C126A mutant of Plasmodium falciparum triosephosphate isomerase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Samanta, M, Banerjee, M, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2010-12-08 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Probing the role of the fully conserved Cys126 in triosephosphate isomerase by site-specific mutagenesis--distal effects on dimer stability.
Febs J., 278, 2011
|
|
3L7F
 
 | | Structure of IL-13 antibody H2L6, A humanized variant OF C836 | | Descriptor: | CALCIUM ION, H2L6 HEAVY CHAIN, H2L6 LIGHT CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-28 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
3S3U
 
 | |
3PRO
 
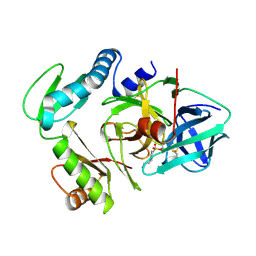 | | ALPHA-LYTIC PROTEASE COMPLEXED WITH C-TERMINAL TRUNCATED PRO REGION | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, ALPHA-LYTIC PROTEASE | | Authors: | Sauter, N.K, Mau, T, Rader, S.D, Agard, D.A. | | Deposit date: | 1998-08-26 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of alpha-lytic protease complexed with its pro region.
Nat.Struct.Biol., 5, 1998
|
|
5YWZ
 
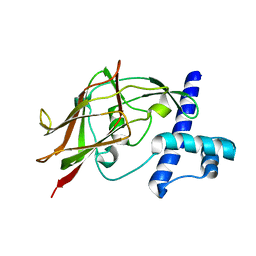 | | AID-SUN tandem of SUN1 | | Descriptor: | SUN domain-containing protein 1 | | Authors: | Xu, Y, Li, W, Feng, W. | | Deposit date: | 2017-11-30 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural conservation of the autoinhibitory domain in SUN proteins
Biochem. Biophys. Res. Commun., 496, 2018
|
|
4AD9
 
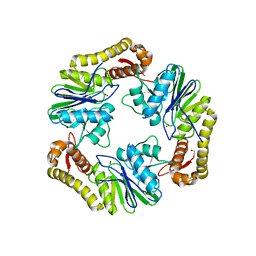 | | Crystal structure of human LACTB2. | | Descriptor: | 1,2-ETHANEDIOL, BETA-LACTAMASE-LIKE PROTEIN 2, ZINC ION | | Authors: | Allerston, C.K, Krojer, T, Shrestha, B, Burgess Brown, N, Chalk, R, Elkins, J.M, Filippakopoulos, P, Pike, A.C.W, Muniz, J.R.C, Vollmar, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, von Delft, F, Gileadi, O. | | Deposit date: | 2011-12-22 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of Lactb2, a Metallo-Beta-Lactamase Protein, as a Human Mitochondrial Endoribonuclease
Nucleic Acids Res., 44, 2016
|
|
3PVF
 
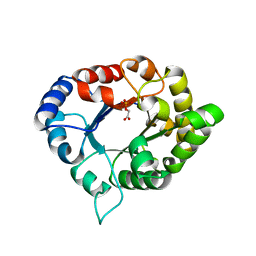 | | Structure of C126S mutant of Plasmodium falciparum triosephosphate isomerase complexed with PGA | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Samanta, M, Banerjee, M, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2010-12-07 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Probing the role of the fully conserved Cys126 in triosephosphate isomerase by site-specific mutagenesis--distal effects on dimer stability.
Febs J., 278, 2011
|
|
2BBF
 
 | | Crystal structure of tRNA-guanine transglycosylase (TGT) from Zymomonas mobilis in complex with 6-amino-3,7-dihydro-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-AMINO-3,7-DIHYDRO-IMIDAZO[4,5-G]QUINAZOLIN-8-ONE, ZINC ION, tRNA guanine transglycosylase | | Authors: | Stengl, B, Meyer, E.A, Heine, A, Brenk, R, Diederich, F, Klebe, G. | | Deposit date: | 2005-10-17 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of tRNA-guanine transglycosylase (TGT) in complex with novel and potent inhibitors unravel pronounced induced-fit adaptations and suggest dimer formation upon substrate binding.
J.Mol.Biol., 370, 2007
|
|
3LTJ
 
 | | Structure of a new family of artificial alpha helicoidal repeat proteins (alpha-Rep) based on thermostable HEAT-like repeats | | Descriptor: | AlphaRep-4 | | Authors: | Urvoas, A, Guellouz, A, Graille, M, van Tilbeurgh, H, Desmadril, M, Minard, P. | | Deposit date: | 2010-02-16 | | Release date: | 2010-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, production and molecular structure of a new family of artificial alpha-helicoidal repeat proteins ( alpha Rep) based on thermostable HEAT-like repeats
J.Mol.Biol., 404, 2010
|
|
1J2T
 
 | | Creatininase Mn | | Descriptor: | MANGANESE (II) ION, SULFATE ION, ZINC ION, ... | | Authors: | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | Deposit date: | 2003-01-11 | | Release date: | 2004-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
2SLI
 
 | | LEECH INTRAMOLECULAR TRANS-SIALIDASE COMPLEXED WITH 2,7-ANHYDRO-NEU5AC, THE REACTION PRODUCT | | Descriptor: | 2-ACETYLAMINO-7-(1,2-DIHYDROXY-ETHYL)-3-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCTANE-5-CARBOXYLIC ACID, INTRAMOLECULAR TRANS-SIALIDASE | | Authors: | Luo, Y, Li, S.C, Li, Y.T, Luo, M. | | Deposit date: | 1998-10-03 | | Release date: | 1999-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A structures of leech intramolecular trans-sialidase complexes: evidence of its enzymatic mechanism.
J.Mol.Biol., 285, 1999
|
|
1ZXK
 
 | | Crystal Structure of Cadherin8 EC1 domain | | Descriptor: | Cadherin-8 | | Authors: | Patel, S.D, Ciatto, C, Chen, C.P, Bahna, F, Arkus, N, Schieren, I, Rajebhosale, M, Jessell, T.M, Honig, B, Price, S.R, Shapiro, L. | | Deposit date: | 2005-06-08 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Type II cadherin ectodomain structures: implications for classical cadherin specificity.
Cell(Cambridge,Mass.), 124, 2006
|
|
2K35
 
 | | Hydramacin-1: Structure and antibacterial activity of a peptide from the basal metazoan Hydra | | Descriptor: | hydramacin-1 | | Authors: | Jung, S, Dingley, A.J, Stanisak, M, Gelhaus, C, Bosch, T, Podschun, R, Leippe, M, Gr tzinger, J. | | Deposit date: | 2008-04-22 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Hydramacin-1, structure and antibacterial activity of a protein from the Basal metazoan hydra.
J.Biol.Chem., 284, 2009
|
|
2MDV
 
 | |
8XF1
 
 | | Crystal structure of the dissociated C-phycocyanin beta-chain from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Nguyen, H.K, Teramoto, T, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Disassembly and reassembly of the non-conventional thermophilic C-phycocyanin.
J.Biosci.Bioeng., 137, 2024
|
|
8XF0
 
 | | Crystal structure of the dissociated C-phycocyanin alpha-chain from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin alpha chain, MAGNESIUM ION, PHYCOCYANOBILIN | | Authors: | Nguyen, H.K, Teramoto, T, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Disassembly and reassembly of the non-conventional thermophilic C-phycocyanin.
J.Biosci.Bioeng., 137, 2024
|
|
