2P8E
 
 | | Crystal structure of the serine/threonine phosphatase domain of human PPM1B | | Descriptor: | MAGNESIUM ION, PPM1B beta isoform variant 6 | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Lau, C, Xu, W, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-22 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2OQV
 
 | | Human Dipeptidyl Peptidase IV (DPP4) with piperidine-constrained phenethylamine | | Descriptor: | (3R,4R)-1-{6-[3-(METHYLSULFONYL)PHENYL]PYRIMIDIN-4-YL}-4-(2,4,5-TRIFLUOROPHENYL)PIPERIDIN-3-AMINE, Dipeptidyl peptidase 4 (Dipeptidyl peptidase IV) (DPP IV) | | Authors: | Pei, Z, Li, X, von Geldern, T.W, Longenecker, K.L, Pireh, D, Stewart, K.D. | | Deposit date: | 2007-02-01 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Structure-Activity Relationships of Piperidinone- and Piperidine-Constrained Phenethylamines as Novel, Potent, and Selective Dipeptidyl Peptidase IV Inhibitors.
J.Med.Chem., 50, 2007
|
|
2P2H
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a pyridinyl-triazine inhibitor | | Descriptor: | 4-(2-anilinopyridin-3-yl)-N-(3,4,5-trimethoxyphenyl)-1,3,5-triazin-2-amine, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Kim, J.L, Long, A.M, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2007-03-07 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Evolution of a Highly Selective and Potent 2-(Pyridin-2-yl)-1,3,5-triazine Tie-2 Kinase Inhibitor
J.Med.Chem., 50, 2007
|
|
2P50
 
 | | Crystal structure of N-acetyl-D-Glucosamine-6-Phosphate deacetylase liganded with Zn | | Descriptor: | N-acetylglucosamine-6-phosphate deacetylase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Hall, R.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2007-03-14 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Diversity within the Mononuclear and Binuclear Active Sites of N-Acetyl-d-glucosamine-6-phosphate Deacetylase.
Biochemistry, 46, 2007
|
|
2QCU
 
 | | Crystal structure of Glycerol-3-phosphate Dehydrogenase from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, Aerobic glycerol-3-phosphate dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yeh, J.I, Chinte, U, Du, S. | | Deposit date: | 2007-06-19 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of glycerol-3-phosphate dehydrogenase, an essential monotopic membrane enzyme involved in respiration and metabolism.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2QEJ
 
 | | Crystal structure of a Staphylococcus aureus protein (SSL7) in complex with Fc of human IgA1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Ramsland, P.A, Willoughby, N, Trist, H.M, Farrugia, W, Hogarth, P.M, Fraser, J.D, Wines, B.D. | | Deposit date: | 2007-06-26 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for evasion of IgA immunity by Staphylococcus aureus revealed in the complex of SSL7 with Fc of human IgA1
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2QOE
 
 | | Human Dipeptidyl Peptidase IV in complex with a Triazolopiperazine-based beta amino acid Inhibitor | | Descriptor: | (2R)-4-[(8R)-8-METHYL-2-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[1,5-A]PYRAZIN-7(8H)-YL]-4-OXO-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-AMINE, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2007-07-20 | | Release date: | 2007-11-06 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, synthesis, and biological evaluation of triazolopiperazine-based beta-amino amides as potent, orally active dipeptidyl peptidase IV (DPP-4) inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2R0B
 
 | | Crystal structure of human tyrosine phosphatase-like serine/threonine/tyrosine-interacting protein | | Descriptor: | GLYCEROL, SULFATE ION, Serine/threonine/tyrosine-interacting protein | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Iizuka, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-18 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2QS4
 
 | | Crystal structure of the GluR5 ligand binding core dimer in complex with LY466195 at 1.58 Angstroms resolution | | Descriptor: | (3S,4aR,6S,8aR)-6-{[(2S)-2-carboxy-4,4-difluoropyrrolidin-1-yl]methyl}decahydroisoquinoline-3-carboxylic acid, AMMONIUM ION, GLYCEROL, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
2QO3
 
 | | Crystal Structure of [KS3][AT3] didomain from module 3 of 6-deoxyerthronolide B synthase | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, ACETATE ION, ... | | Authors: | Khosla, C, Cane, E.D, Tang, Y, Chen, Y.A, Kim, C.Y. | | Deposit date: | 2007-07-19 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural and mechanistic analysis of protein interactions in module 3 of the 6-deoxyerythronolide B synthase
Chem.Biol., 14, 2007
|
|
2QUO
 
 | |
2QM8
 
 | | MeaB, A Bacterial Homolog of MMAA, in the Nucleotide Free Form | | Descriptor: | GTPase/ATPase, PHOSPHATE ION | | Authors: | Hubbard, P.A, Padovani, D, Labunska, T, Mahlstedt, S.A, Banerjee, R, Drennan, C.L. | | Deposit date: | 2007-07-14 | | Release date: | 2007-08-28 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and mutagenesis of the metallochaperone MeaB: insight into the causes of methylmalonic aciduria.
J.Biol.Chem., 282, 2007
|
|
2QOM
 
 | | The crystal structure of the E.coli EspP autotransporter Beta-domain. | | Descriptor: | Serine protease espP | | Authors: | Barnard, T.J, Dautin, N, Lukacik, P, Bernstein, H.D, Buchanan, S.K. | | Deposit date: | 2007-07-20 | | Release date: | 2007-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Autotransporter structure reveals intra-barrel cleavage followed by conformational changes.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2QT9
 
 | | Human dipeptidyl peptidase iv/cd26 in complex with a 4-aryl cyclohexylalanine inhibitor | | Descriptor: | (2S,3S)-3-AMINO-4-[(3S)-3-FLUOROPYRROLIDIN-1-YL]-N,N-DIMETHYL-4-OXO-2-(TRANS-4-[1,2,4]TRIAZOLO[1,5-A]PYRIDIN-5-YLCYCLOH EXYL)BUTANAMIDE, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2007-08-01 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 4-Arylcyclohexylalanine analogs as potent, selective, and orally active inhibitors of dipeptidyl peptidase IV.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2R1U
 
 | |
2R4J
 
 | | Crystal structure of Escherichia coli SeMet substituted Glycerol-3-phosphate Dehydrogenase in complex with DHAP | | Descriptor: | 1,2-ETHANEDIOL, 1,3-DIHYDROXYACETONEPHOSPHATE, Aerobic glycerol-3-phosphate dehydrogenase, ... | | Authors: | Yeh, J.I, Du, S, Chinte, U. | | Deposit date: | 2007-08-31 | | Release date: | 2008-06-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of glycerol-3-phosphate dehydrogenase, an essential monotopic membrane enzyme involved in respiration and metabolism.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2QBW
 
 | | The crystal structure of PDZ-Fibronectin fusion protein | | Descriptor: | PDZ-Fibronectin fusion protein, Polypeptide | | Authors: | Huang, J, Makabe, K, Koide, A, Koide, S. | | Deposit date: | 2007-06-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design of protein function leaps by directed domain interface evolution.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3KWF
 
 | | human DPP-IV with carmegliptin (S)-1-((2S,3S,11bS)-2-Amino-9,10-dimethoxy-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-3-yl)-4-fluoromethyl-pyrrolidin-2-one | | Descriptor: | (4S)-1-[(2S,3S,11bS)-2-amino-9,10-dimethoxy-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-3-yl]-4-(fluoromethyl)pyrrolidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 | | Authors: | Hennig, M, Stihle, M, Thoma, R. | | Deposit date: | 2009-12-01 | | Release date: | 2010-01-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of carmegliptin: A potent and long-acting dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4OC4
 
 | | X-ray structure of of human glutamate carboxypeptidase II (GCPII) in a complex with CPIBzL, a urea-based inhibitor N~2~-{[(1S)-1-carboxy-2-(pyridin-4-yl)ethyl]carbamoyl}-N~6~-(4-iodobenzoyl)-L-lysine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Pavlicek, J, Ptacek, J, Cerny, J, Byun, Y, Skultetyova, L, Pomper, M, Lubkowski, J, Barinka, C. | | Deposit date: | 2014-01-08 | | Release date: | 2014-05-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural characterization of P1'-diversified urea-based inhibitors of glutamate carboxypeptidase II.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3KM8
 
 | |
4ORL
 
 | |
3KYF
 
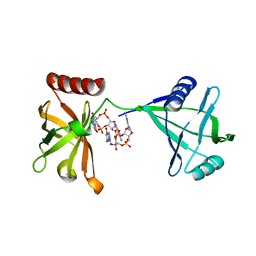 | | Crystal structure of P4397 complexed with c-di-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Putative uncharacterized protein | | Authors: | Ryu, K.S, Ko, J, Kim, H, Choi, B.S. | | Deposit date: | 2009-12-06 | | Release date: | 2010-04-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of PP4397 Reveals the Molecular Basis for Different c-di-GMP Binding Modes by Pilz Domain Proteins.
J.Mol.Biol., 398, 2010
|
|
4OBI
 
 | |
4OGF
 
 | |
4OGZ
 
 | |
