7E7X
 
 | |
7EK6
 
 | | Structure of viral peptides IPB19/N52 | | Descriptor: | Spike protein S2 | | Authors: | Yu, D, Qin, B, Cui, S, He, Y. | | Deposit date: | 2021-04-04 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.243 Å) | | Cite: | Structure-based design and characterization of novel fusion-inhibitory lipopeptides against SARS-CoV-2 and emerging variants.
Emerg Microbes Infect, 10, 2021
|
|
7DZX
 
 | | Spike protein from SARS-CoV2 with Fab fragment of enhancing antibody 8D2 | | Descriptor: | Fab Heavy chain of enhancing antibody, Fab light chain of enhancing antibody, Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
7E88
 
 | |
7E8C
 
 | | SARS-CoV-2 S-6P in complex with 9 Fabs | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E8F
 
 | | SARS-CoV-2 NTD in complex with N9 Fab | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E86
 
 | |
7EB0
 
 | | Cryo-EM structure of SARS-CoV-2 Spike D614G variant, one RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-23 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation.
J.Biol.Chem., 297, 2021
|
|
7EJ4
 
 | | Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody RBD-chAb-25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RBD-chAb-25, ... | | Authors: | Yang, T.J, Yu, P.Y, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-04-01 | | Release date: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody RBD-chAb-25
To be published
|
|
7EAZ
 
 | | Cryo-EM structure of SARS-CoV-2 Spike D614G variant, one RBD-up conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-23 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation.
J.Biol.Chem., 297, 2021
|
|
7EJ5
 
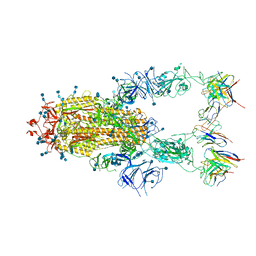 | | Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody RBD-chAb-45 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RBD-chAb45, ... | | Authors: | Yang, T.J, Yu, P.Y, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-04-01 | | Release date: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody RBD-chAb-45
To Be Published
|
|
7E5Y
 
 | | Molecular basis for neutralizing antibody 2B11 targeting SARS-CoV-2 RBD | | Descriptor: | 2B11 Fab Heavy chain, 2B11 Fab Light chain, Spike protein S1 | | Authors: | Wu, H, Yu, F, Wang, Q.S, Zhou, H, Wang, W.W, Zhao, T, Pan, Y.B, Yang, X.M. | | Deposit date: | 2021-02-21 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Screening of potent neutralizing antibodies against SARS-CoV-2 using convalescent patients-derived phage-display libraries.
Cell Discov, 7, 2021
|
|
7EB5
 
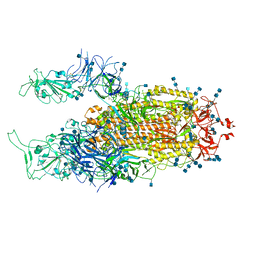 | | Cryo-EM structure of SARS-CoV-2 Spike D614G variant, two RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-23 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation.
J.Biol.Chem., 297, 2021
|
|
7EB3
 
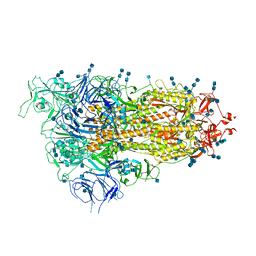 | | Cryo-EM structure of SARS-CoV-2 Spike D614G variant, one RBD-up conformation 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-23 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation.
J.Biol.Chem., 297, 2021
|
|
7JJI
 
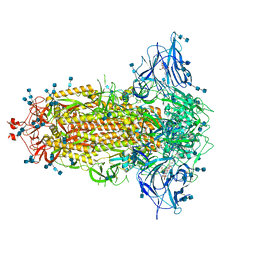 | | Structure of SARS-CoV-2 3Q-2P full-length prefusion spike trimer (C3 symmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-hydroxyethyl 2-deoxy-3,5-bis-O-(2-hydroxyethyl)-6-O-(2-{[(9E)-octadec-9-enoyl]oxy}ethyl)-alpha-L-xylo-hexofuranoside, ... | | Authors: | Bangaru, S, Turner, H.L, Ozorowski, G, Antanasijevic, A, Ward, A.B. | | Deposit date: | 2020-07-26 | | Release date: | 2020-08-26 | | Last modified: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate.
Science, 370, 2020
|
|
7JJJ
 
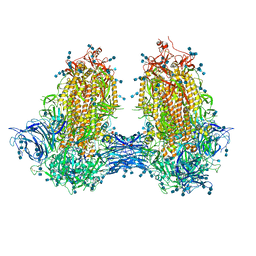 | | Structure of SARS-CoV-2 3Q-2P full-length dimers of spike trimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bangaru, S, Turner, H.L, Ozorowski, G, Antanasijevic, A, Ward, A.B. | | Deposit date: | 2020-07-26 | | Release date: | 2020-08-26 | | Last modified: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate.
Science, 370, 2020
|
|
7EAM
 
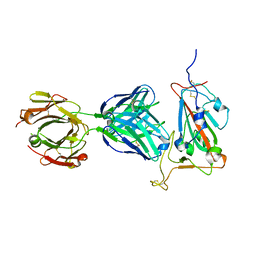 | | immune complex of SARS-CoV-2 RBD and cross-neutralizing antibody 7D6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Li, T.T, Gu, Y, Li, S.W. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cross-neutralizing antibodies bind a SARS-CoV-2 cryptic site and resist circulating variants.
Nat Commun, 12, 2021
|
|
7DWY
 
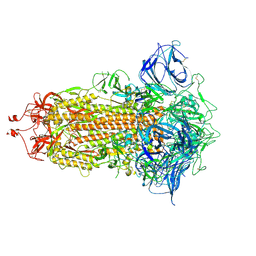 | | S protein of SARS-CoV-2 in the locked conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DX3
 
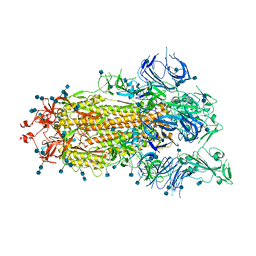 | | S protein of SARS-CoV-2 bound with PD of ACE2 in the conformation 1 (1 up RBD and no PD bound) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DX0
 
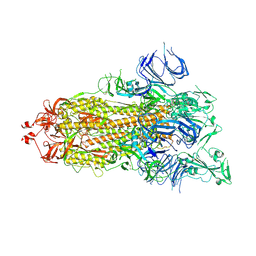 | | Trypsin-digested S protein of SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DWZ
 
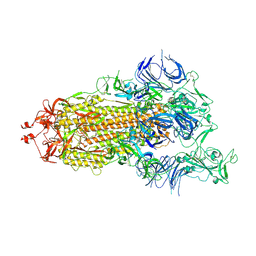 | | S protein of SARS-CoV-2 in the active conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DX1
 
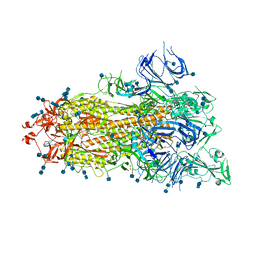 | | S protein of SARS-CoV-2 D614G mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DX2
 
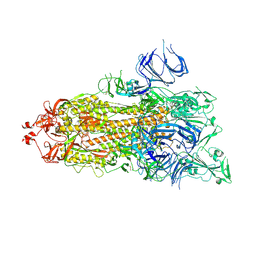 | | Trypsin-digested S protein of SARS-CoV-2 D614G mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7EAN
 
 | | immune complex of SARS-CoV-2 RBD and cross-neutralizing antibody 6D6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of SARS-CoV-2 cross-neutralizing mAb 6D6, Light chain of SARS-CoV-2 cross-neutralizing mAb 6D6, ... | | Authors: | Li, T.T, Gu, Y, Li, S.W. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Cross-neutralizing antibodies bind a SARS-CoV-2 cryptic site and resist circulating variants.
Nat Commun, 12, 2021
|
|
7E3J
 
 | |
