8TK5
 
 | | HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE (VHR) complexed with HEPES | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Aleshin, A.E, Wu, J, Lambert, L.J, Cosford, N.D.P, Tautz, L. | | Deposit date: | 2023-07-25 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment Screening Platform and Discovery of Novel Fragment Binders of the VHR Phosphatase, a Drug Target for Sepsis and Septic Shock
To Be Published
|
|
8TK4
 
 | | HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE (VHR) complexed with phosphate | | Descriptor: | Dual specificity protein phosphatase 3, PHOSPHATE ION | | Authors: | Aleshin, A.E, Wu, J, Lambert, L.J, Cosford, N.D.P, Tautz, L. | | Deposit date: | 2023-07-25 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE (VHR) complexed with phosphate
To Be Published
|
|
5Z59
 
 | | Crystal structure of Tk-PTP in the inactive form | | Descriptor: | Protein-tyrosine phosphatase | | Authors: | Ku, B, Yun, H.Y, Kim, S.J. | | Deposit date: | 2018-01-17 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Structural study reveals the temperature-dependent conformational flexibility of Tk-PTP, a protein tyrosine phosphatase from Thermococcus kodakaraensis KOD1
PLoS ONE, 13, 2018
|
|
5Z5B
 
 | | Crystal structure of Tk-PTP in the G95A mutant form | | Descriptor: | CHLORIDE ION, FORMIC ACID, Protein-tyrosine phosphatase | | Authors: | Ku, B, Yun, H.Y, Kim, S.J. | | Deposit date: | 2018-01-17 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural study reveals the temperature-dependent conformational flexibility of Tk-PTP, a protein tyrosine phosphatase from Thermococcus kodakaraensis KOD1
PLoS ONE, 13, 2018
|
|
5XJV
 
 | | Two intermediate states of conformation switch in dual specificity phosphatase 13a | | Descriptor: | Dual specificity protein phosphatase 13 isoform A, PHOSPHATE ION | | Authors: | Wei, C.H, Min, H.G, Chun, H.J, Ryu, S.E. | | Deposit date: | 2017-05-04 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Two intermediate states of the conformational switch in dual specificity phosphatase 13a
Pharmacol. Res., 128, 2018
|
|
5Z5A
 
 | | Crystal structure of Tk-PTP in the active form | | Descriptor: | Protein-tyrosine phosphatase, VANADATE ION | | Authors: | Ku, B, Yun, H.Y, Kim, S.J. | | Deposit date: | 2018-01-17 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural study reveals the temperature-dependent conformational flexibility of Tk-PTP, a protein tyrosine phosphatase from Thermococcus kodakaraensis KOD1
PLoS ONE, 13, 2018
|
|
6L1S
 
 | | Crystal structure of DUSP22 mutant_C88S | | Descriptor: | Dual specificity protein phosphatase 22, PHOSPHATE ION | | Authors: | Lai, C.H, Chang, C.C, Lyu, P.C. | | Deposit date: | 2019-09-30 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3611 Å) | | Cite: | Structural Insights into the Active Site Formation of DUSP22 in N-loop-containing Protein Tyrosine Phosphatases.
Int J Mol Sci, 21, 2020
|
|
6LVQ
 
 | | Crystal structure of DUSP22_VO4 | | Descriptor: | Dual specificity protein phosphatase 22, VANADATE ION | | Authors: | Lai, C.H, Lyu, P.C. | | Deposit date: | 2020-02-04 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural Insights into the Active Site Formation of DUSP22 in N-loop-containing Protein Tyrosine Phosphatases.
Int J Mol Sci, 21, 2020
|
|
6LOU
 
 | | Crystal structure of DUSP22 mutant_C88S/S93N | | Descriptor: | Dual specificity protein phosphatase 22, PHOSPHATE ION | | Authors: | Lai, C.H, Lyu, P.C. | | Deposit date: | 2020-01-07 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5301 Å) | | Cite: | Structural Insights into the Active Site Formation of DUSP22 in N-loop-containing Protein Tyrosine Phosphatases.
Int J Mol Sci, 21, 2020
|
|
6MC1
 
 | | Structure of MAP kinase phosphatase 5 in complex with 3,3-dimethyl-1-((9-(methylthio)-5,6-dihydrothieno[3,4-h]quinazolin-2-yl)thio)butan-2-one, an allosteric inhibitor | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3,3-dimethyl-1-{[9-(methylsulfanyl)-5,6-dihydrothieno[3,4-h]quinazolin-2-yl]sulfanyl}butan-2-one, ACETATE ION, ... | | Authors: | Gannam, Z.T.K, Anderson, K.S, Bennett, A.M, Lolis, E. | | Deposit date: | 2018-08-30 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An allosteric site on MKP5 reveals a strategy for small-molecule inhibition.
Sci.Signal., 13, 2020
|
|
6LMY
 
 | | Crystal structure of DUSP22 mutant_C88S/S93A | | Descriptor: | Dual specificity protein phosphatase 22, PHOSPHATE ION | | Authors: | Lai, C.H, Lyu, P.C. | | Deposit date: | 2019-12-27 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights into the Active Site Formation of DUSP22 in N-loop-containing Protein Tyrosine Phosphatases.
Int J Mol Sci, 21, 2020
|
|
6LOT
 
 | | Crystal structure of DUSP22 mutant_N128D | | Descriptor: | Dual specificity protein phosphatase 22, SULFATE ION | | Authors: | Lai, C.H, Lyu, P.C. | | Deposit date: | 2020-01-07 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural Insights into the Active Site Formation of DUSP22 in N-loop-containing Protein Tyrosine Phosphatases.
Int J Mol Sci, 21, 2020
|
|
3NME
 
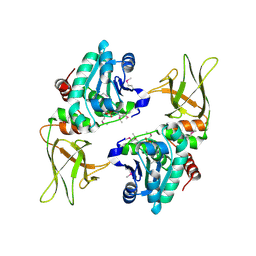 | | Structure of a plant phosphatase | | Descriptor: | PHOSPHATE ION, SEX4 glucan phosphatase | | Authors: | Vander Kooi, C.W. | | Deposit date: | 2010-06-22 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the glucan phosphatase activity of Starch Excess4.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6APX
 
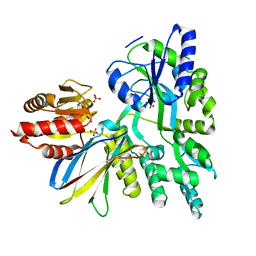 | | Crystal structure of human dual specificity phosphatase 1 catalytic domain (C258S) as a maltose binding protein fusion in complex with the monobody YSX1 | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein,Dual specificity protein phosphatase 1, Monobody YSX1, ... | | Authors: | Gumpena, R, Lountos, G.T, Sreejith, R.K, Tropea, J.E, Cherry, S, Waugh, D.S. | | Deposit date: | 2017-08-18 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Crystal structure of the human dual specificity phosphatase 1 catalytic domain.
Protein Sci., 27, 2018
|
|
6D65
 
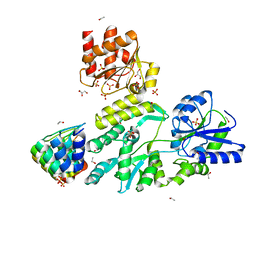 | | Crystal structure of the human dual specificity phosphatase 1 catalytic domain (C258S) as a maltose binding protein fusion in complex with the designed AR protein off7 | | Descriptor: | Designed AR protein off7, ETHANOL, GLYCEROL, ... | | Authors: | Gumpena, R, Lountos, G.T, Waugh, D.S. | | Deposit date: | 2018-04-20 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | MBP-binding DARPins facilitate the crystallization of an MBP fusion protein.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
4PYH
 
 | |
6D66
 
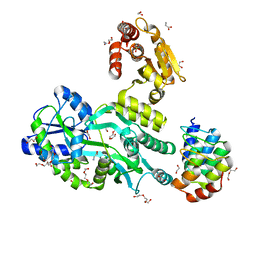 | | Crystal structure of the human dual specificity 1 catalytic domain (C258S) as a maltose binding protein fusion in complex with the designed AR protein mbp3_16 | | Descriptor: | 1,2-ETHANEDIOL, D-ALANINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gumpena, R, Waugh, D.S, Lountos, G.T. | | Deposit date: | 2018-04-20 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | MBP-binding DARPins facilitate the crystallization of an MBP fusion protein.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6D67
 
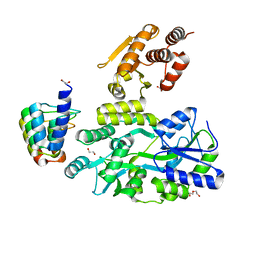 | | Crystal structure of the human dual specificity phosphatase 1 catalytic domain (C258S) as a maltose binding protein fusion (maltose bound form) in complex with the designed AR protein mbp3_16 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Designed AR protein mbp3_16, ... | | Authors: | Gumpena, R, Lountos, G.T, Waugh, D.S. | | Deposit date: | 2018-04-20 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | MBP-binding DARPins facilitate the crystallization of an MBP fusion protein.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6G84
 
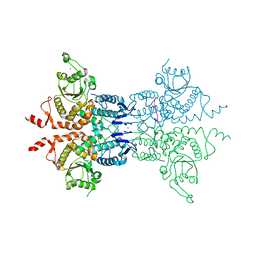 | | Structure of Cdc14 bound to CBK1 PxL motif | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CBK1, ... | | Authors: | Mouilleron, S, Kataria, M, Uhlmann, F. | | Deposit date: | 2018-04-07 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | A PxL motif promotes timely cell cycle substrate dephosphorylation by the Cdc14 phosphatase.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6G86
 
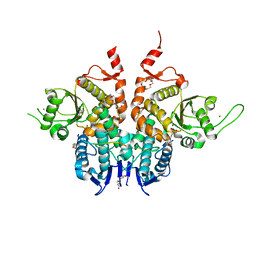 | | Structure of Cdc14 bound to SIC1 PxL motif | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, ... | | Authors: | Mouilleron, S, Kataria, M, Uhlmann, F. | | Deposit date: | 2018-04-07 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A PxL motif promotes timely cell cycle substrate dephosphorylation by the Cdc14 phosphatase.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6G85
 
 | | Structure of Cdc14 bound to CBK1 PxL motif | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CBK1, ... | | Authors: | Mouilleron, S, Kataria, M, Uhlmann, F. | | Deposit date: | 2018-04-07 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.528 Å) | | Cite: | A PxL motif promotes timely cell cycle substrate dephosphorylation by the Cdc14 phosphatase.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5XW4
 
 | |
5XW5
 
 | |
6RZZ
 
 | |
4YR8
 
 | | Crystal structure of JNK in complex with a regulator protein | | Descriptor: | CHLORIDE ION, Dual specificity protein phosphatase 16, Mitogen-activated protein kinase 8 | | Authors: | Liu, X, Wang, J, Wu, J.W, Wang, Z.X. | | Deposit date: | 2015-03-14 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A conserved motif in JNK/p38-specific MAPK phosphatases as a determinant for JNK1 recognition and inactivation.
Nat Commun, 7, 2016
|
|
