6OA1
 
 | | Structure of human PARG complexed with JA2120 | | Descriptor: | 1,3-dimethyl-8-{[2-(morpholin-4-yl)ethyl]sulfanyl}-3,7-dihydro-1H-purine-2,6-dione, Poly(ADP-ribose) glycohydrolase | | Authors: | Stegeman, R.A, Jones, D.E, Ellenberger, T, Kim, I.K, Tainer, J.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
7PX6
 
 | | PARP15 catalytic domain in complex with OUL241 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-[(2-fluorophenyl)methoxy]phthalazine-1,4-dione, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2021-10-08 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Potent 2,3-dihydrophthalazine-1,4-dione derivatives as dual inhibitors for mono-ADP-ribosyltransferases PARP10 and PARP15.
Eur.J.Med.Chem., 237, 2022
|
|
7PWQ
 
 | | PARP15 catalytic domain in complex with OUL240 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-(cyclohexylmethoxy)phthalazine-1,4-dione, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2021-10-07 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Potent 2,3-dihydrophthalazine-1,4-dione derivatives as dual inhibitors for mono-ADP-ribosyltransferases PARP10 and PARP15.
Eur.J.Med.Chem., 237, 2022
|
|
7PX7
 
 | | PARP15 catalytic domain in complex with OUL242 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-(thiophen-2-ylmethoxy)phthalazine-1,4-dione, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2021-10-08 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Potent 2,3-dihydrophthalazine-1,4-dione derivatives as dual inhibitors for mono-ADP-ribosyltransferases PARP10 and PARP15.
Eur.J.Med.Chem., 237, 2022
|
|
7Q0V
 
 | | Lysozyme soaked with V(IV)OSO4 and phen | | Descriptor: | 1,10-PHENANTHROLINE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Santos, M.F.A, Fernandes, A.C.P, Correia, I, Sciortino, G, Garribba, E, Santos-Silva, T, Pessoa, J.C. | | Deposit date: | 2021-10-16 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Binding of V IV O 2+ , V IV OL, V IV OL 2 and V V O 2 L Moieties to Proteins: X-ray/Theoretical Characterization and Biological Implications.
Chemistry, 28, 2022
|
|
7PZG
 
 | | Phocaeicola vulgatus sialic acid esterase at 1.44 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, Lysophospholipase L1, MAGNESIUM ION, ... | | Authors: | Scott, H, Armstrong, Z, Davies, G.J. | | Deposit date: | 2021-10-12 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | The structure of Phocaeicola vulgatus sialic acid acetylesterase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PZ7
 
 | | Structure of an LPMO at 1.13x10^6 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACRYLIC ACID, ... | | Authors: | Tandrup, T, Muderspach, S.J, Ipsen, J.O, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
8R8P
 
 | |
6PGE
 
 | |
9DE7
 
 | | Structure of full-length HIV TAR RNA G16A/A17G | | Descriptor: | RNA (58-MER) | | Authors: | Bou-Nader, C, Zhang, J. | | Deposit date: | 2024-08-28 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of complete HIV-1 TAR RNA portray a dynamic platform poised for protein binding and structural remodeling.
Nat Commun, 16, 2025
|
|
6P58
 
 | | Dark and Steady State-Illuminated Crystal Structure of Cyanobacteriochrome Receptor PixJ at 150K | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Clinger, J.A, Miller, M.D, Buirgie, E.S, Vierstra, R.D, Phillips Jr, G.N. | | Deposit date: | 2019-05-29 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4W79
 
 | | Crystal Structure of Human Protein N-terminal Glutamine Amidohydrolase | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, Protein N-terminal glutamine amidohydrolase, ... | | Authors: | Bitto, E, Bingman, C.A, McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human protein N-terminal glutamine amidohydrolase, an initial component of the N-end rule pathway.
Plos One, 9, 2014
|
|
9R9E
 
 | | Recombinant human butyrylcholinesterase in complex with N-(3-methoxypropyl)-N-{[1-(prop-2-yn-1-yl)pyrrolidin-3-yl]methyl}naphthalene-2-sulfonamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Brazzolotto, X, Kosak, U, Nachon, F, Gobec, S. | | Deposit date: | 2025-05-20 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | N-Propargylpyrrolidine-based butyrylcholinesterase and monoamine oxidase inhibitors.
Chem.Biol.Interact., 420, 2025
|
|
4UTR
 
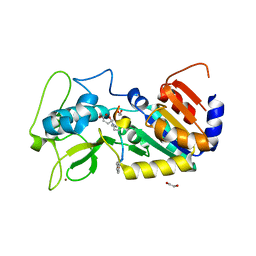 | | Crystal structure of zebrafish Sirtuin 5 in complex with glutarylated CPS1-peptide | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, GLUTARIC ACID, ... | | Authors: | Pannek, M, Gertz, M, Steegborn, C. | | Deposit date: | 2014-07-22 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Chemical Probing of the Human Sirtuin 5 Active Site Reveals its Substrate Acyl Specificity and Peptide-Based Inhibitors.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4UV6
 
 | |
9IZV
 
 | | Cryo-EM structure of ALDH6A1-Y172H & R535C | | Descriptor: | Methylmalonate-semialdehyde/malonate-semialdehyde dehydrogenase [acylating], mitochondrial | | Authors: | Su, G, Xu, Y, Luan, X. | | Deposit date: | 2024-08-01 | | Release date: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural and biochemical basis for the pathogenic mutations of methylmalonate semialdehyde dehydrogenase ALDH6A1
Medicine Plus, 1, 2024
|
|
9IZX
 
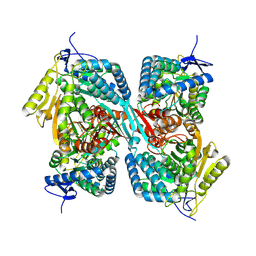 | | Cryo-EM structure of ALDH6A1-G446R | | Descriptor: | Methylmalonate-semialdehyde/malonate-semialdehyde dehydrogenase [acylating], mitochondrial | | Authors: | Su, G, Xu, Y, Luan, X. | | Deposit date: | 2024-08-01 | | Release date: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and biochemical basis for the pathogenic mutations of methylmalonate semialdehyde dehydrogenase ALDH6A1
Medicine Plus, 1, 2024
|
|
8CKW
 
 | |
8CKV
 
 | |
8CL1
 
 | |
8CL0
 
 | |
4W9Q
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-5-ethyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-5-ethyl-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Pomplun, S, Wang, Y, Kirschner, K, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Rational Design and Asymmetric Synthesis of Potent and Neurotrophic Ligands for FK506-Binding Proteins (FKBPs).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
8CKX
 
 | |
6VO5
 
 | | Crystal structure of Human histone acetytransferas 1 (HAT1) in complex with isobutryl-COA and K12A mutant variant of histone H4 | | Descriptor: | ACETATE ION, GLYCEROL, Histone H4, ... | | Authors: | Halabelian, L, Zeng, H, Dong, A, Loppnau, P, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-01-30 | | Release date: | 2020-03-11 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Human histone acetytransferas 1 (HAT1) in complex with isobutryl-COA and K12A mutant variant of histone H4
to be published
|
|
8CCZ
 
 | |
