1SM7
 
 | | Solution structure of the recombinant pronapin precursor, BnIb. | | Descriptor: | recombinant Ib pronapin | | Authors: | Pantoja-Uceda, D, Palomares, O, Bruix, M, Villalba, M, Rodriguez, R, Rico, M, Santoro, J. | | Deposit date: | 2004-03-08 | | Release date: | 2005-02-01 | | Last modified: | 2013-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and stability against digestion of rproBnIb, a recombinant 2S albumin from rapeseed: relationship to its allergenic properties.
Biochemistry, 43, 2004
|
|
1FYC
 
 | | INNER LIPOYL DOMAIN FROM HUMAN PYRUVATE DEHYDROGENASE (PDH) COMPLEX, NMR, 1 STRUCTURE | | Descriptor: | DIHYDROLIPOAMIDE ACETYLTRANSFERASE (E2P) | | Authors: | Howard, M.J, Fuller, C, Broadhurst, R.W, Quinn, J, Yeaman, S.J, Perham, R.N. | | Deposit date: | 1997-02-21 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the major autoantigen in primary biliary cirrhosis.
Gastroenterology, 115, 1998
|
|
5FEG
 
 | |
3TC6
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR63. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Indole-3-glycerol phosphate synthase, PHOSPHATE ION | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Exploration of alternate catalytic mechanisms and optimization strategies for retroaldolase design.
J.Mol.Biol., 426, 2014
|
|
1K34
 
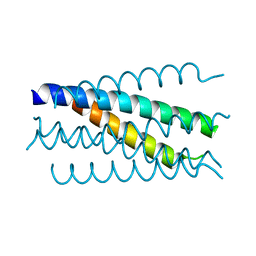 | | Crystal structure analysis of gp41 core mutant | | Descriptor: | Transmembrane glycoprotein GP41 | | Authors: | Shu, W, Lu, M. | | Deposit date: | 2001-10-01 | | Release date: | 2001-10-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Interhelical interactions in the gp41 core: implications for activation of HIV-1 membrane fusion.
Biochemistry, 41, 2002
|
|
3TC7
 
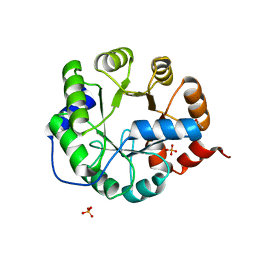 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR62. | | Descriptor: | ACETIC ACID, Indole-3-glycerol phosphate synthase, PHOSPHATE ION | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exploration of alternate catalytic mechanisms and optimization strategies for retroaldolase design.
J.Mol.Biol., 426, 2014
|
|
4AH2
 
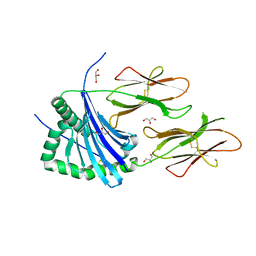 | | HLA-DR1 with covalently linked CLIP106-120 in canonical orientation | | Descriptor: | GLYCEROL, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN GAMMA CHAIN, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN,DRB1-1 BETA CHAIN, ... | | Authors: | Schlundt, A, Guenther, S, Sticht, J, Wieczorek, M, Roske, Y, Heinemann, U, Freund, C. | | Deposit date: | 2012-02-03 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Peptide Linkage to the Alpha-Subunit of Mhcii Creates a Stably Inverted Antigen Presentation Complex.
J.Mol.Biol., 423, 2012
|
|
4AGI
 
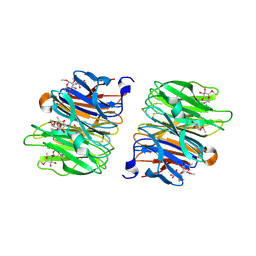 | | Crystal Structure of Fucose binding lectin from Aspergillus Fumigatus (AFL) in complex with seleno fucoside. | | Descriptor: | FUCOSE-SPECIFIC LECTIN FLEA, methyl 1-seleno-alpha-L-fucopyranoside | | Authors: | Houser, J, Komarek, J, Kostlanova, N, Lahmann, M, Cioci, G, Varrot, A, Imberty, A, Wimmerova, M. | | Deposit date: | 2012-01-30 | | Release date: | 2013-02-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Soluble Fucose-Specific Lectin from Aspergillus Fumigatus Conidia - Structure, Specificity and Possible Role in Fungal Pathogenicity.
Plos One, 8, 2013
|
|
3V0R
 
 | | Crystal structure of Alternaria alternata allergen Alt a 1 | | Descriptor: | 2,5,6-triaminopyrimidin-4-ol, 8-aminooctanoic acid, Major allergen Alt a 1, ... | | Authors: | Chruszcz, M, Solberg, R, Osinski, T, Chapman, M.D, Minor, W. | | Deposit date: | 2011-12-08 | | Release date: | 2012-06-13 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alternaria alternata allergen Alt a 1: a unique beta-barrel protein dimer found exclusively in fungi.
J.Allergy Clin.Immunol., 130, 2012
|
|
5HHV
 
 | |
5HI3
 
 | |
5FEF
 
 | |
5FDS
 
 | |
5X4R
 
 | | Structure of the N-terminal domain (NTD) of MERS-CoV spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X58
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
6IM7
 
 | | CueO-12.1 multicopper oxidase | | Descriptor: | Blue copper oxidase CueO,12.1 peptide,Blue copper oxidase CueO, CALCIUM ION | | Authors: | Wongsantichon, J, Robinson, R, Ghadessy, F. | | Deposit date: | 2018-10-22 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Development and structural characterization of an engineered multi-copper oxidase reporter of protein-protein interactions.
J.Biol.Chem., 294, 2019
|
|
3RRC
 
 | |
5GNB
 
 | | Crystal Structure of the Receptor Binding Domain of the Spike Glycoprotein of Human Betacoronavirus HKU1 (HKU1 1A-CTD, 2.3 angstrom, native-SAD phasing) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Guan, H, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2016-07-20 | | Release date: | 2017-06-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the receptor binding domain of the spike glycoprotein of human betacoronavirus HKU1
Nat Commun, 8, 2017
|
|
6IM9
 
 | | MDM2 bound CueO-PM2 sensor | | Descriptor: | Blue copper oxidase CueO,PM2 peptide,Blue copper oxidase CueO, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Wongsantichon, J, Robinson, R, Ghadessy, F. | | Deposit date: | 2018-10-22 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Development and structural characterization of an engineered multi-copper oxidase reporter of protein-protein interactions.
J.Biol.Chem., 294, 2019
|
|
4Q7D
 
 | | Wild type Fc (wtFc) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region | | Authors: | Ahmed, A.A, Giddens, J, Pincetic, A, Lomino, J.V, Ravetch, J.V, Wang, L.X, Bjorkman, P.J. | | Deposit date: | 2014-04-24 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization of anti-inflammatory immunoglobulin g fc proteins.
J.Mol.Biol., 426, 2014
|
|
5X4S
 
 | | Structure of the N-terminal domain (NTD)of SARS-CoV spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
3UL9
 
 | | structure of the TV3 mutant M41E | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Toll-like receptor 4, ... | | Authors: | Kim, H.J, Cheong, H.K, Jeon, Y.H. | | Deposit date: | 2011-11-10 | | Release date: | 2012-05-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Rational Design of a Toll-like Receptor 4 (TLR4) Decoy Receptor with High Binding Affinity for a Target Protein.
Plos One, 7, 2012
|
|
5HHC
 
 | | Crystal Structure of Chemically Synthesized Heterochiral {RFX037 plus VEGF-A} Protein Complex in space group P21/n | | Descriptor: | D- Vascular endothelial growth factor-A, GLYCEROL, Vascular endothelial growth factor A | | Authors: | Uppalapati, M, Lee, D.J, Mandal, K, Kent, S.B.H, Sidhu, S. | | Deposit date: | 2016-01-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Potent d-Protein Antagonist of VEGF-A is Nonimmunogenic, Metabolically Stable, and Longer-Circulating in Vivo.
Acs Chem.Biol., 11, 2016
|
|
1GX0
 
 | | ALPHA-,1,3 GALACTOSYLTRANSFERASE - BETA-D-GALACTOSE COMPLEX | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Boix, E, Zhang, Y, Swaminathan, G.J, Brew, K, Acharya, K.R. | | Deposit date: | 2002-03-26 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Ordered Binding of Donor and Acceptor Substrates to the Retaining Glycosyltransferase, Alpha -1,3 Galactosyltransferase
J.Biol.Chem., 277, 2002
|
|
1IUD
 
 | | MALTODEXTRIN-BINDING PROTEIN INSERTION/DELETION MUTANT WITH AN INSERTED B-CELL EPITOPE FROM THE PRES2 REGION OF HEPATITIS B VIRUS | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN MALE-B133, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Saul, F.A, Vulliez-Le Normand, B, Lema, F, Bentley, G.A. | | Deposit date: | 1996-05-29 | | Release date: | 1997-06-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a recombinant form of the maltodextrin-binding protein carrying an inserted sequence of a B-cell epitope from the preS2 region of hepatitis B virus.
Proteins, 27, 1997
|
|
