6EE4
 
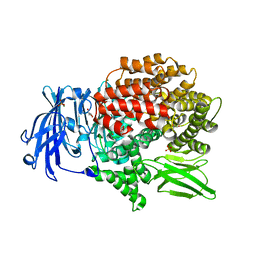 | | X-ray crystal structure of Pf-M1 in complex with inhibitor (6m) and catalytic zinc ion | | Descriptor: | (2R)-2-[(cyclopropylacetyl)amino]-N-hydroxy-2-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)acetamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2018-08-13 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Hydroxamic Acid Inhibitors Provide Cross-Species Inhibition of Plasmodium M1 and M17 Aminopeptidases.
J. Med. Chem., 62, 2019
|
|
6U7G
 
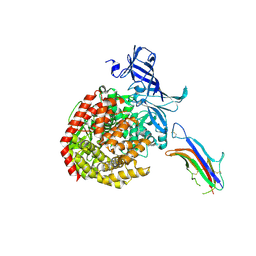 | | HCoV-229E RBD Class V in complex with human APN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Tomlinson, A, Li, Z, Rini, J.M. | | Deposit date: | 2019-09-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The human coronavirus HCoV-229E S-protein structure and receptor binding.
Elife, 8, 2019
|
|
6U7F
 
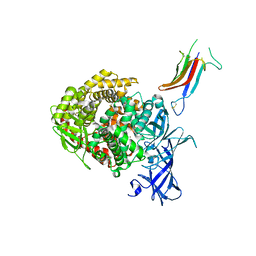 | | HCoV-229E RBD Class IV in complex with human APN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Tomlinson, A.C.A, Li, Z, Rini, J.M. | | Deposit date: | 2019-09-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The human coronavirus HCoV-229E S-protein structure and receptor binding.
Elife, 8, 2019
|
|
6EE6
 
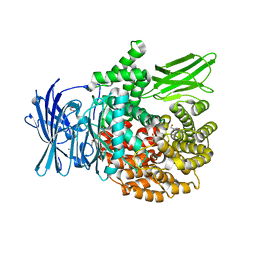 | | X-ray crystal structure of Pf-M1 in complex with inhibitor (6o) and catalytic zinc ion | | Descriptor: | (2R)-2-[(cyclopentylacetyl)amino]-N-hydroxy-2-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)acetamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2018-08-13 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hydroxamic Acid Inhibitors Provide Cross-Species Inhibition of Plasmodium M1 and M17 Aminopeptidases.
J. Med. Chem., 62, 2019
|
|
5Y28
 
 | | Crystal structure of H. pylori HtrA with PDZ2 deletion | | Descriptor: | 1,2-ETHANEDIOL, Periplasmic serine endoprotease DegP-like, UNK-UNK-UNK-UNK | | Authors: | Zhang, Z, Huang, Q, Tao, X. | | Deposit date: | 2017-07-24 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.08606815 Å) | | Cite: | The unique trimeric assembly of the virulence factor HtrA fromHelicobacter pylorioccurs via N-terminal domain swapping.
J.Biol.Chem., 294, 2019
|
|
4DWU
 
 | |
5Y2D
 
 | | Crystal structure of H. pylori HtrA | | Descriptor: | Periplasmic serine endoprotease DegP-like, UNK-UNK-K-UNK-UNK-UNK-UNK-UNK-UNK-UNK, UNK-UNK-UNK, ... | | Authors: | Zhang, Z, Huang, Q, Tao, X. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.70009851 Å) | | Cite: | The unique trimeric assembly of the virulence factor HtrA fromHelicobacter pylorioccurs via N-terminal domain swapping.
J.Biol.Chem., 294, 2019
|
|
4DWT
 
 | |
1A16
 
 | | AMINOPEPTIDASE P FROM E. COLI WITH THE INHIBITOR PRO-LEU | | Descriptor: | AMINOPEPTIDASE P, LEUCINE, MANGANESE (II) ION, ... | | Authors: | Wilce, M.C, Bond, C.S, Lilley, P.E, Dixon, N.E, Freeman, H.C, Guss, J.M. | | Deposit date: | 1997-12-22 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of a proline-specific aminopeptidase from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
4BH9
 
 | |
6EHI
 
 | | NucT from Helicobacter pylori | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Celma, L, Li de la Sierra-Gallay, I, Quevillon-Cheruel, S. | | Deposit date: | 2017-09-13 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural basis for the substrate selectivity of Helicobacter pylori NucT nuclease activity.
PLoS ONE, 12, 2017
|
|
4V6V
 
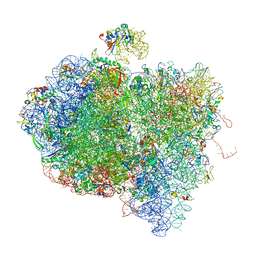 | | Tetracycline resistance protein Tet(O) bound to the ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Li, W, Atkinson, G.C, Thakor, N.S, Allas, U, Lu, C, Chan, K.Y, Tenson, T, Schulten, K, Wilson, K.S, Hauryliuk, V, Frank, J. | | Deposit date: | 2013-02-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Mechanism of tetracycline resistance by ribosomal protection protein Tet(O).
Nat Commun, 4, 2013
|
|
4AJF
 
 | | Identification and structural characterization of PDE10 fragment inhibitors | | Descriptor: | CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, MAGNESIUM ION, ... | | Authors: | Johansson, P, Albert, J.S, Spadola, L, Akerud, T, Back, E, Hillertz, P, Horsefeld, R, Scott, C, Spear, N, Tian, G, Tigerstrom, A, Aharony, D, Geschwindner, S. | | Deposit date: | 2012-02-16 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and Structural Characterization of Pde10 Fragment Inhibitors
To be Published
|
|
4X2U
 
 | | X-ray crystal structure of the orally available aminopeptidase inhibitor, Tosedostat, bound to the M1 Alanyl Aminopeptidase from P. falciparum | | Descriptor: | (2S)-({(2R)-2-[(1S)-1-hydroxy-2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}amino)(phenyl)ethanoic acid, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2014-11-27 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystal structures of an orally available aminopeptidase inhibitor, Tosedostat, bound to anti-malarial drug targets PfA-M1 and PfA-M17.
Proteins, 83, 2015
|
|
4X8I
 
 | | de novo crystal structure of the Pyrococcus Furiosus TET3 aminopeptidase | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Colombo, M. | | Deposit date: | 2014-12-10 | | Release date: | 2016-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tuned by metals: the TET peptidase activity is controlled by 3 metal binding sites.
Sci Rep, 6, 2016
|
|
4XO4
 
 | |
4XN8
 
 | |
1B6A
 
 | | HUMAN METHIONINE AMINOPEPTIDASE 2 COMPLEXED WITH TNP-470 | | Descriptor: | (1R,2S,3S,4R)-4-hydroxy-2-methoxy-4-methyl-3-[(2R,3R)-2-methyl-3-(3-methylbut-2-en-1-yl)oxiran-2-yl]cyclohexyl (chloroacetyl)carbamate, COBALT (II) ION, METHIONINE AMINOPEPTIDASE | | Authors: | Liu, S, Clardy, J.C. | | Deposit date: | 1999-01-13 | | Release date: | 2000-01-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of human methionine aminopeptidase-2 complexed with fumagillin.
Science, 282, 1998
|
|
4XN2
 
 | |
4XN4
 
 | |
4XN7
 
 | |
1B59
 
 | | COMPLEX OF HUMAN METHIONINE AMINOPEPTIDASE-2 COMPLEXED WITH OVALICIN | | Descriptor: | 3,4-DIHYDROXY-2-METHOXY-4-METHYL-3-[2-METHYL-3-(3-METHYL-BUT-2-ENYL) -OXIRANYL]-CYCLOHEXANONE, COBALT (II) ION, PROTEIN (METHIONINE AMINOPEPTIDASE) | | Authors: | Liu, S, Clardy, J.C. | | Deposit date: | 1999-01-13 | | Release date: | 2000-01-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human methionine aminopeptidase-2 complexed with fumagillin.
Science, 282, 1998
|
|
4XND
 
 | |
4XMU
 
 | |
4XMZ
 
 | |
