5XF6
 
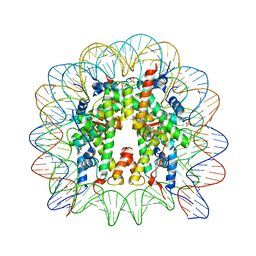 | | Nucleosome core particle with an adduct of a binuclear RAPTA (Ru-arene-phosphaadamantane) compound having an ethylenediamine linker | | Descriptor: | DNA (145-MER), ETHANE-1,2-DIAMINE, Histone H2A, ... | | Authors: | Ma, Z, Adhireksan, Z, Murray, B.S, Dyson, P.J, Davey, C.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Nucleosome acidic patch-targeting binuclear ruthenium compounds induce aberrant chromatin condensation
Nat Commun, 8, 2017
|
|
6UG3
 
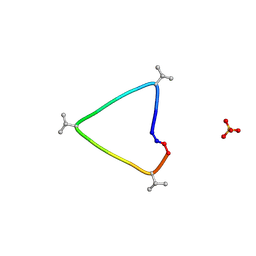 | | C3 symmetric peptide design number 1, Sporty, crystal form 1 | | Descriptor: | C3-1, Sporty, crystal form 1, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
5TUS
 
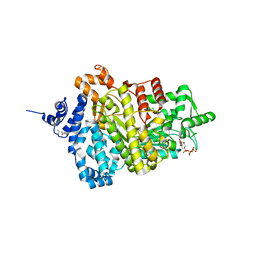 | | Potent competitive inhibition of human ribonucleotide reductase by a novel non-nucleoside small molecule | | Descriptor: | 2-hydroxy-N'-[(Z)-(2-hydroxynaphthalen-1-yl)methylidene]benzohydrazide, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | Authors: | Mohammed, F.A, Alam, I, Dealwis, C.G. | | Deposit date: | 2016-11-07 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Potent competitive inhibition of human ribonucleotide reductase by a nonnucleoside small molecule.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8BXY
 
 | | FimH in complex with alpha1,6 core-fucosylated oligomannose-3, crystallized in the trigonal space group | | Descriptor: | NICKEL (II) ION, SULFATE ION, Type 1 fimbrin D-mannose specific adhesin, ... | | Authors: | Bridot, C, Bouckaert, J, Krammer, E.-M. | | Deposit date: | 2022-12-11 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into a cooperative switch between one and two FimH bacterial adhesins binding pauci- and high-mannose type N-glycan receptors.
J.Biol.Chem., 299, 2023
|
|
7C5F
 
 | | Crystal Structure of Glyceraldehyde-3-phosphate dehydrogenase1 from Escherichia coli at 1.88 Angstrom resolution | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Zhang, L, Liu, M.R, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Characterization and structure of glyceraldehyde-3-phosphate dehydrogenase type 1 from Escherichia coli.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
8C78
 
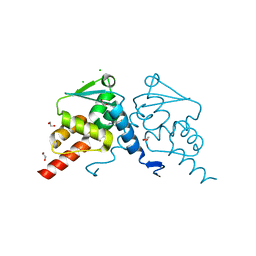 | | Crystal structure of human BCL6 BTB domain in complex with compound CCT374705 | | Descriptor: | (2~{S})-10-[(3-chloranyl-2-fluoranyl-pyridin-4-yl)amino]-2-cyclopropyl-3,3-bis(fluoranyl)-7-methyl-2,4-dihydro-1~{H}-[1,4]oxazepino[2,3-c]quinolin-6-one, 1,2-ETHANEDIOL, B-cell lymphoma 6 protein, ... | | Authors: | Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2023-01-13 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of an In Vivo Chemical Probe for BCL6 Inhibition by Optimization of Tricyclic Quinolinones.
J.Med.Chem., 66, 2023
|
|
1TOA
 
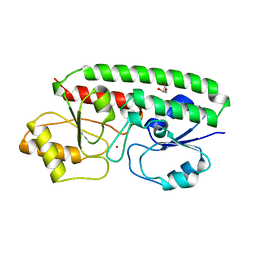 | | PERIPLASMIC ZINC BINDING PROTEIN TROA FROM TREPONEMA PALLIDUM | | Descriptor: | GLYCEROL, PROTEIN (PERIPLASMIC BINDING PROTEIN TROA), ZINC ION | | Authors: | Lee, Y.H, Deka, R.K, Norgard, M.V, Radolf, J.D, Hasemann, C.A. | | Deposit date: | 1999-03-03 | | Release date: | 1999-07-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Treponema pallidum TroA is a periplasmic zinc-binding protein with a helical backbone.
Nat.Struct.Biol., 6, 1999
|
|
1A63
 
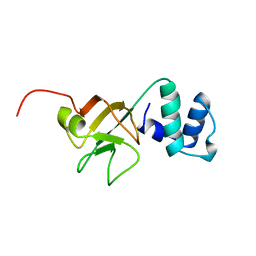 | | THE NMR STRUCTURE OF THE RNA BINDING DOMAIN OF E.COLI RHO FACTOR SUGGESTS POSSIBLE RNA-PROTEIN INTERACTIONS, 10 STRUCTURES | | Descriptor: | RHO | | Authors: | Briercheck, D.M, Wood, T.C, Allison, T.J, Richardson, J.P, Rule, G.S. | | Deposit date: | 1998-03-05 | | Release date: | 1998-05-27 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the RNA binding domain of E. coli rho factor suggests possible RNA-protein interactions.
Nat.Struct.Biol., 5, 1998
|
|
6KH4
 
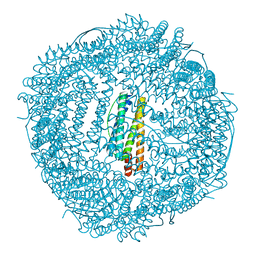 | | Design and crystal structure of protein MOFs with ferritin nanocages as linkers and nickel clusters as nodes | | Descriptor: | FE (III) ION, Ferritin, NICKEL (II) ION | | Authors: | Gu, C, Chen, H, Wang, Y, Zhang, T, Wang, H, Zhao, G. | | Deposit date: | 2019-07-12 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural Insight into Binary Protein Metal-Organic Frameworks with Ferritin Nanocages as Linkers and Nickel Clusters as Nodes.
Chemistry, 26, 2020
|
|
6JU1
 
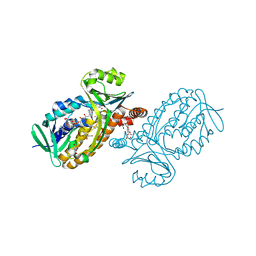 | | p-Hydroxybenzoate hydroxylase Y385F mutant complexed with 3,4-dihydroxybenzoate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 4-hydroxybenzoate 3-monooxygenase, ... | | Authors: | Yato, M, Arakawa, T, Yamada, C, Fushinobu, S. | | Deposit date: | 2019-04-12 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Understanding the Molecular Mechanism Underlying the High Catalytic Activity ofp-Hydroxybenzoate Hydroxylase Mutants for Producing Gallic Acid.
Biochemistry, 58, 2019
|
|
4YXP
 
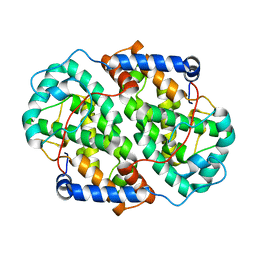 | | The structure of the folded domain of the signature multifunctional protein ICP27 from herpes simplex virus-1 reveals an intertwined dimer. | | Descriptor: | ZINC ION, mRNA export factor | | Authors: | Tunnicliffe, R.B, Schacht, M, Levy, C.W, Jowitt, T.A, Sandri-Goldin, R.M, Golovanov, A.P. | | Deposit date: | 2015-03-23 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The structure of the folded domain from the signature multifunctional protein ICP27 from herpes simplex virus-1 reveals an intertwined dimer.
Sci Rep, 5, 2015
|
|
1BBA
 
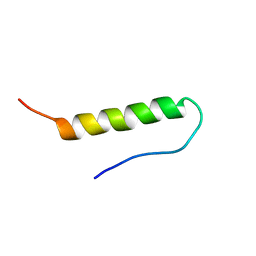 | |
6QJ4
 
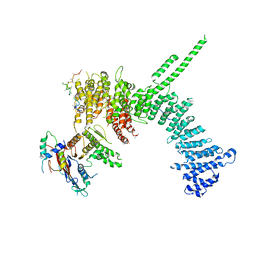 | | Crystal structure of the C. thermophilum condensin Ycs4-Brn1 subcomplex bound to the Smc4 ATPase head in complex with the C-terminal domain of Brn1 | | Descriptor: | Brn1, Condensin complex subunit 1,Condensin complex subunit 1,Condensin complex subunit 1, Condensin complex subunit 2, ... | | Authors: | Hassler, M, Haering, C.H, Kschonsak, M. | | Deposit date: | 2019-01-22 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (5.8 Å) | | Cite: | Structural Basis of an Asymmetric Condensin ATPase Cycle.
Mol.Cell, 74, 2019
|
|
6B2X
 
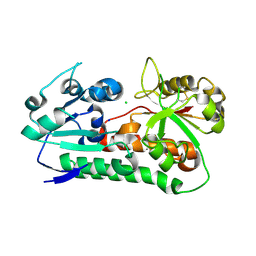 | | Apo YiuA Crystal Form 1 | | Descriptor: | CHLORIDE ION, SODIUM ION, Solute-binding periplasmic protein of iron/siderophore ABC transporter | | Authors: | Radka, C.D, DeLucas, L.J, Aller, S.G. | | Deposit date: | 2017-09-20 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | The crystal structure of the Yersinia pestis iron chaperone YiuA reveals a basic triad binding motif for the chelated metal.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1V0M
 
 | | Xylanase Xyn10a from Streptomyces lividans in complex with xylobio-deoxynojirimycin at pH 7.5 | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, IMIDAZOLE, PIPERIDINE-3,4,5-TRIOL, ... | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
8ZLH
 
 | | The crystal structure of CcmS. | | Descriptor: | All1292 protein | | Authors: | Cheng, J, Li, C.L. | | Deposit date: | 2024-05-20 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular interactions of the chaperone CcmS and carboxysome shell protein CcmK1 that mediate beta-carboxysome assembly.
Plant Physiol., 196, 2024
|
|
1AGF
 
 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGKKRYKL-5R MUTATION) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGKKRYKL - 5R MUTATION) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
4X2W
 
 | |
7QYG
 
 | | Structure of the transaminase TR2 | | Descriptor: | Aminotransferase TR2 | | Authors: | Roda, S, Fernandez-Lopez, L, Benedens, M, Bollinger, A, Thies, S, Schumacher, J, Coscolin, C, Kazemi, M, Santiago, G, Gertzen, C.G, Gonzalez-Alfonso, J, Plou, F.J, Jaeger, K.E, Smits, S.H, Ferrer, M, Guallar, V. | | Deposit date: | 2022-01-28 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A Plurizyme with Transaminase and Hydrolase Activity Catalyzes Cascade Reactions.
Angew Chem Int Ed Engl, 61, 2022
|
|
4X6Q
 
 | | An Isoform-specific Myristylation Switch Targets RIIb PKA Holoenzymes to Membranes | | Descriptor: | cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase type II-beta regulatory subunit | | Authors: | Zhang, P, Ye, F, Bastidas, A.C, Kornev, A.P, Ginsberg, M.H, Taylor, S.S. | | Deposit date: | 2014-12-08 | | Release date: | 2015-07-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | An Isoform-Specific Myristylation Switch Targets Type II PKA Holoenzymes to Membranes.
Structure, 23, 2015
|
|
1AGD
 
 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGKKKYKL-INDEX PEPTIDE) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGKKKYKL - INDEX PEPTIDE) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
4R3E
 
 | |
1URO
 
 | | UROPORPHYRINOGEN DECARBOXYLASE | | Descriptor: | BETA-MERCAPTOETHANOL, PROTEIN (UROPORPHYRINOGEN DECARBOXYLASE) | | Authors: | Whitby, F.G, Phillips, J.D, Kushner, J.P, Hill, C.P. | | Deposit date: | 1998-08-21 | | Release date: | 1998-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human uroporphyrinogen decarboxylase.
EMBO J., 17, 1998
|
|
6JOG
 
 | | Crystal structure of the complex of phospho pantetheine adenylyl transferase from Acinetobacter baumannii with two ascorbic acid (Vitamin-C) molecules. | | Descriptor: | ASCORBIC ACID, Phosphopantetheine adenylyltransferase, SULFATE ION | | Authors: | Viswanathan, V, Gupta, A, Bairagya, H.R, Sharma, S, Singh, T.P. | | Deposit date: | 2019-03-20 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the complex of phospho pantetheine adenylyl transferase from Acinetobacter baumannii with two ascorbic acid (Vitamin-C) molecules.
To Be Published
|
|
9DMM
 
 | | Crystal structure of human KRAS G12C covalently bound to Divarasib (GDC6036) | | Descriptor: | 1-{(3S)-4-[(7M)-7-[6-amino-4-methyl-3-(trifluoromethyl)pyridin-2-yl]-6-chloro-8-fluoro-2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}quinazolin-4-yl]-3-methylpiperazin-1-yl}propan-1-one, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Fernando, M.C. | | Deposit date: | 2024-09-13 | | Release date: | 2025-05-21 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of KRAS G12C bound to divarasib highlights features of potent switch-II pocket engagement.
Small Gtpases, 15, 2024
|
|
