1UXX
 
 | | CBM6ct from Clostridium thermocellum in complex with xylopentaose | | Descriptor: | CALCIUM ION, XYLANASE U, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Henrissat, D.B.B, Gilbert, H.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
1UYY
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with cellotriose | | Descriptor: | CALCIUM ION, CELLULASE B, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
5MAP
 
 | | X-ray generated oxyferrous complex of DtpA from Streptomyces lividans | | Descriptor: | DtpA, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Moreno Chicano, T, Chaplin, A.K, Worrall, J.A.R, Strange, R.W, Hough, M.A. | | Deposit date: | 2016-11-04 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Photoreduction and validation of haem-ligand intermediate states in protein crystals by in situ single-crystal spectroscopy and diffraction.
IUCrJ, 4, 2017
|
|
4N38
 
 | | Structure of langerin CRD I313 D288 complexed with GlcNAc-beta1-3Gal-beta1-4GlcNAc-beta-CH2CH2N3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, C-type lectin domain family 4 member K, CALCIUM ION, ... | | Authors: | Feinberg, H, Rowntree, T.J.W, Tan, S.L.W, Drickamer, K, Weis, W.I, Taylor, M.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Common polymorphisms in human langerin change specificity for glycan ligands.
J.Biol.Chem., 288, 2013
|
|
4N35
 
 | | Structure of langerin CRD I313 complexed with GlcNAc-beta1-3Gal-beta1-4Glc-beta-CH2CH2N3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, C-type lectin domain family 4 member K, ... | | Authors: | Feinberg, H, Rowntree, T.J.W, Tan, S.L.W, Drickamer, K, Weis, W.I, Taylor, M.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-11-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Common polymorphisms in human langerin change specificity for glycan ligands.
J.Biol.Chem., 288, 2013
|
|
2O5R
 
 | |
1FQB
 
 | | STRUCTURE OF MALTOTRIOTOL BOUND TO OPEN-FORM MALTODEXTRIN BINDING PROTEIN IN P2(1)CRYSTAL FORM | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-sorbitol | | Authors: | Duan, X, Hall, J.A, Nikaido, H, Quiocho, F.A. | | Deposit date: | 2000-09-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: flexibility of tertiary structure and ligand binding.
J.Mol.Biol., 306, 2001
|
|
1PXJ
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR 4-(2,4-Dimethyl-thiazol-5-yl)-pyrimidin-2-ylamine | | Descriptor: | 4-(2,4-DIMETHYL-1,3-THIAZOL-5-YL)PYRIMIDIN-2-AMINE, Cell division protein kinase 2 | | Authors: | Wu, S.Y, McNae, I, Kontopidis, G, McClue, S.J, McInnes, C, Stewart, K.J, Wang, S, Zheleva, D.I, Marriage, H, Lane, D.P, Taylor, P, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2003-07-04 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a novel family of CDK inhibitors with the program LIDAEUS: structural basis for ligand-induced disordering of the activation loop
Structure, 11, 2003
|
|
4OXQ
 
 | | Structure of Staphylococcus pseudintermedius metal-binding protein SitA in complex with Zinc | | Descriptor: | Manganese ABC transporter, periplasmic-binding protein SitA, ZINC ION | | Authors: | Abate, F, Malito, E, Bottomley, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Apo, Zn2+-bound and Mn2+-bound structures reveal ligand-binding properties of SitA from the pathogen Staphylococcus pseudintermedius.
Biosci.Rep., 34, 2014
|
|
3M63
 
 | |
3ZM6
 
 | | CRYSTAL STRUCTURE OF MURF LIGASE IN COMPLEX WITH CYANOTHIOPHENE INHIBITOR | | Descriptor: | N-(6-(4-(2h-tetrazol-5-yl)benzyl)-3-cyano-4,5,6,7-tetrahydrothieno[2,3-c]pyridin-2-yl)-2,4-dichloro-5-(morpholinosulfonyl)benzamide, UDP-N-ACETYLMURAMOYL-TRIPEPTIDE--D-ALANYL-D-ALANINE LIGASE | | Authors: | Hrast, M, Turk, S, Sosic, I, Knez, D, Randall, C.P, Barreteau, H, Contreras-Martel, C, Dessen, A, ONeill, A.J, Mengin-Lecreulx, D, Blanot, D, Gobec, S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-Activity Relationships of New Cyanothiophene Inhibitors of the Essential Peptidoglycan Biosynthesis Enzyme Murf.
Eur.J.Med.Chem., 66C, 2013
|
|
1PO1
 
 | | POLIOVIRUS (TYPE 1, MAHONEY) IN COMPLEX WITH R80633, AN INHIBITOR OF VIRAL REPLICATION | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE BUTYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 1 MAHONEY | | Authors: | Hiremath, C.N, Filman, D.J, Grant, R.A, Hogle, J.M. | | Deposit date: | 1997-01-08 | | Release date: | 1997-12-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand-induced conformational changes in poliovirus-antiviral drug complexes.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
4POW
 
 | | Structure of the PBP NocT in complex with pyronopaline | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1S)-4-carbamimidamido-1-carboxybutyl]-5-oxo-D-proline, Nopaline-binding periplasmic protein | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2014-02-26 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Agrobacterium uses a unique ligand-binding mode for trapping opines and acquiring a competitive advantage in the niche construction on plant host.
Plos Pathog., 10, 2014
|
|
3FLL
 
 | | Crystal structure of E55Q mutant of nitrophorin 4 | | Descriptor: | AMMONIA, Nitrophorin-4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Montfort, W.R, Weichsel, A. | | Deposit date: | 2008-12-18 | | Release date: | 2009-02-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Effect of mutation of carboxyl side-chain amino acids near the heme on the midpoint potentials and ligand binding constants of nitrophorin 2 and its NO, histamine, and imidazole complexes.
J.Am.Chem.Soc., 131, 2009
|
|
1FQA
 
 | | STRUCTURE OF MALTOTETRAITOL BOUND TO OPEN-FORM MALTODEXTRIN BINDING PROTEIN IN P2(1)CRYSTAL FORM | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-sorbitol | | Authors: | Duan, X, Hall, J.A, Nikaido, H, Quiocho, F.A. | | Deposit date: | 2000-09-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: flexibility of tertiary structure and ligand binding.
J.Mol.Biol., 306, 2001
|
|
4M4G
 
 | | Crystal structure of ligand binding domain of CysB, a LysR member from Salmonella typhimurium LT2 in complex with effector ligand, N-acetylserine. | | Descriptor: | DI(HYDROXYETHYL)ETHER, HTH-type transcriptional regulator CysB, N-ACETYL-SERINE | | Authors: | Mittal, M, Singh, A.K, Kumaran, S. | | Deposit date: | 2013-08-07 | | Release date: | 2014-08-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of ligand binding domain of CysB, a LysR member from Salmonella typhimurium LT2 in complex with effector ligand, N-acetylserine
To be Published
|
|
1GJQ
 
 | | Pseudomonas aeruginosa cd1 nitrite reductase reduced cyanide complex | | Descriptor: | CYANIDE ION, HEME C, HEME D, ... | | Authors: | Nurizzo, D, Brown, K, Tegoni, M, Cambillau, C. | | Deposit date: | 2001-08-01 | | Release date: | 2002-08-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cyanide Binding to Cd(1) Nitrite Reductase from Pseudomonas Aeruginosa: Role of the Active-Site His369 in Ligand Stabilization.
Biochem.Biophys.Res.Commun., 291, 2002
|
|
1PO2
 
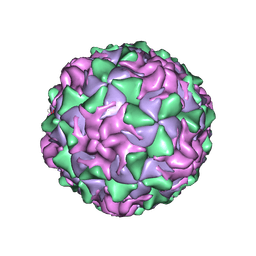 | | POLIOVIRUS (TYPE 1, MAHONEY) IN COMPLEX WITH R77975, AN INHIBITOR OF VIRAL REPLICATION | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE ETHYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 1 MAHONEY | | Authors: | Hiremath, C.N, Filman, D.J, Grant, R.A, Hogle, J.M. | | Deposit date: | 1997-01-08 | | Release date: | 1997-12-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand-induced conformational changes in poliovirus-antiviral drug complexes.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
3ZM5
 
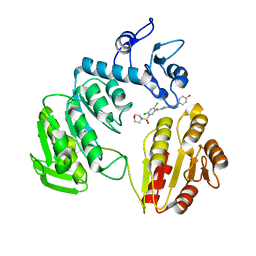 | | CRYSTAL STRUCTURE OF MURF LIGASE IN COMPLEX WITH CYANOTHIOPHENE INHIBITOR | | Descriptor: | 2,4-bis(chloranyl)-N-[3-cyano-6-[(4-hydroxyphenyl)methyl]-5,7-dihydro-4H-thieno[2,3-c]pyridin-2-yl]-5-morpholin-4-ylsulfonyl-benzamide, UDP-N-ACETYLMURAMOYL-TRIPEPTIDE--D-ALANYL-D-ALANINE LIGASE | | Authors: | Hrast, M, Turk, S, Sosic, I, Knez, D, Randall, C.P, Barreteau, H, Contreras-Martel, C, Dessen, A, ONeill, A.J, Mengin-Lecreulx, D, Blanot, D, Gobec, S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structure-Activity Relationships of New Cyanothiophene Inhibitors of the Essential Peptidoglycan Biosynthesis Enzyme Murf.
Eur.J.Med.Chem., 66C, 2013
|
|
1UXZ
 
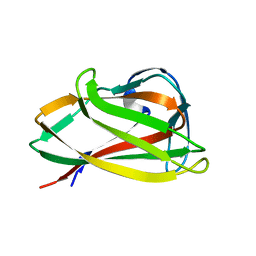 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A | | Descriptor: | CELLULASE B | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
5MJH
 
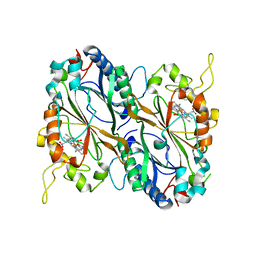 | | X-ray generated oxyferrous/water mixed complex of DtpA from Streptomyces lividans | | Descriptor: | Dye type peroxidase A, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Moreno Chicano, T, Chaplin, A.K, Worrall, J.A.R, Strange, R.W, Hough, M.A. | | Deposit date: | 2016-12-01 | | Release date: | 2017-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Photoreduction and validation of haem-ligand intermediate states in protein crystals by in situ single-crystal spectroscopy and diffraction.
IUCrJ, 4, 2017
|
|
6TFG
 
 | |
6TF1
 
 | |
6TFF
 
 | |
1PXI
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR 4-(2,5-Dichloro-thiophen-3-yl)-pyrimidin-2-ylamine | | Descriptor: | 4-(2,5-DICHLOROTHIEN-3-YL)PYRIMIDIN-2-AMINE, Cell division protein kinase 2 | | Authors: | Wu, S.Y, McNae, I, Kontopidis, G, McClue, S.J, McInnes, C, Stewart, K.J, Wang, S, Zheleva, D.I, Marriage, H, Lane, D.P, Taylor, P, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2003-07-04 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of a novel family of CDK inhibitors with the program LIDAEUS: structural basis for ligand-induced disordering of the activation loop
Structure, 11, 2003
|
|
