1DEW
 
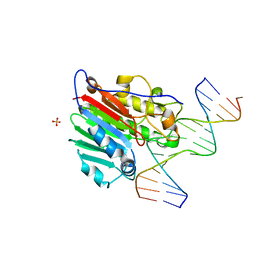 | | CRYSTAL STRUCTURE OF HUMAN APE1 BOUND TO ABASIC DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*(3DR)P*CP*GP*AP*CP*GP*AP*CP*G)-3', 5'-D(*GP*TP*CP*GP*TP*CP*GP*GP*GP*GP*AP*CP*GP*C)-3', MAJOR APURINIC/APYRIMIDINIC ENDONUCLEASE, ... | | Authors: | Mol, C.D, Izumi, T, Mitra, S, Tainer, J.A. | | Deposit date: | 1999-11-15 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination
Nature, 403, 2000
|
|
1DU5
 
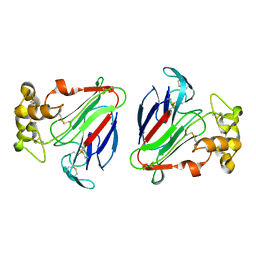 | | THE CRYSTAL STRUCTURE OF ZEAMATIN. | | Descriptor: | ZEAMATIN | | Authors: | Batalia, M.A, Monzingo, A.F, Ernst, S, Roberts, W, Robertus, J.D. | | Deposit date: | 2000-01-14 | | Release date: | 2000-02-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the antifungal protein zeamatin, a member of the thaumatin-like, PR-5 protein family.
Nat.Struct.Biol., 3, 1996
|
|
1QE7
 
 | |
1DV4
 
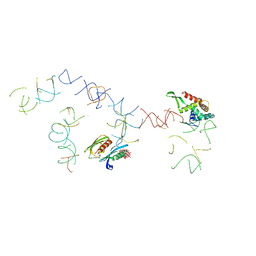 | | PARTIAL STRUCTURE OF 16S RNA OF THE SMALL RIBOSOMAL SUBUNIT FROM THERMUS THERMOPHILUS | | Descriptor: | 16S RIBOSOMAL RNA, OCTADECATUNGSTENYL DIPHOSPHATE, RIBOSOMAL PROTEIN S5, ... | | Authors: | Tocilj, A, Schlunzen, F, Janell, D, Gluhmann, M, Hansen, H, Harms, J, Bashan, A, Bartels, H, Agmon, I, Franceschi, F, Yonath, A. | | Deposit date: | 2000-01-19 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | The small ribosomal subunit from Thermus thermophilus at 4.5 A resolution: pattern fittings and the identification of a functional site.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1DT9
 
 | |
1D6U
 
 | | CRYSTAL STRUCTURE OF E. COLI AMINE OXIDASE ANAEROBICALLY REDUCED WITH BETA-PHENYLETHYLAMINE | | Descriptor: | 2-PHENYLETHYLAMINE, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Wilmot, C.M, Hajdu, J, McPherson, M.J, Knowles, P.F, Phillips, S.E.V. | | Deposit date: | 1999-10-15 | | Release date: | 2000-02-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Visualization of dioxygen bound to copper during enzyme catalysis.
Science, 286, 1999
|
|
1DGQ
 
 | | NMR SOLUTION STRUCTURE OF THE INSERTED DOMAIN OF HUMAN LEUKOCYTE FUNCTION ASSOCIATED ANTIGEN-1 | | Descriptor: | LEUKOCYTE FUNCTION ASSOCIATED ANTIGEN-1 | | Authors: | Legge, G.B, Kriwacki, R.W, Chung, J, Hommel, U, Ramage, P, Case, D.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 1999-11-24 | | Release date: | 2000-02-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the inserted domain of human leukocyte function associated antigen-1.
J.Mol.Biol., 295, 2000
|
|
1QJS
 
 | | mammalian blood serum haemopexin glycosylated-native protein and in complex with its ligand haem | | Descriptor: | CHLORIDE ION, HEMOPEXIN, PHOSPHATE ION, ... | | Authors: | Paoli, M, Baker, H.M, Morgan, W.T, Smith, A, Baker, E.N. | | Deposit date: | 1999-07-01 | | Release date: | 2000-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Hemopexin Reveals a Novel High-Affinity Heme Site Formed between Two Beta-Propeller Domains.
Nat.Struct.Biol., 6, 1999
|
|
1B8Z
 
 | | HU FROM THERMOTOGA MARITIMA | | Descriptor: | PROTEIN (HISTONELIKE PROTEIN HU) | | Authors: | Christodoulou, E, Rypniewski, W.R, Vorgias, C.E. | | Deposit date: | 1999-02-03 | | Release date: | 2000-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cloning, overproduction, purification and crystallization of the DNA binding protein HU from the hyperthermophilic eubacterium Thermotoga maritima.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1QL4
 
 | | Structure of the soluble domain of cytochrome c552 from Paracoccus denitrificans in the oxidised state | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Harrenga, A, Reincke, B, Rueterjans, H, Ludwig, B, Michel, H. | | Deposit date: | 1999-08-20 | | Release date: | 2000-02-03 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Soluble Domain of Cytochrome C552 from Paracoccus Denitrificans in the Oxidized and Reduced States
J.Mol.Biol., 295, 2000
|
|
1QKY
 
 | | Solution structure of PI7, a non toxic peptide isolated from the scorpion Pandinus Imperator. | | Descriptor: | TOXIN 7 FROM PANDINUS IMPERATOR | | Authors: | Delepierre, M, Prochnicka-Chalufour, A, Boisbouvier, J, Possani, L.D. | | Deposit date: | 1999-08-17 | | Release date: | 2000-02-03 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Pi7, an orphan peptide from the scorpion Pandinus imperator: a 1H-NMR analysis using a nano-NMR Probe.
Biochemistry, 38, 1999
|
|
1QMA
 
 | | Nuclear Transport Factor 2 (NTF2) W7A mutant | | Descriptor: | NUCLEAR TRANSPORT FACTOR 2 | | Authors: | Bayliss, R, Ribbeck, K, Akin, D, Kent, H.M, Feldherr, C.M, Gorlich, D, Stewart, M.J. | | Deposit date: | 1999-09-23 | | Release date: | 2000-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interaction Betweeen Ntf2 and Xfxfg-Containing Nucleoporins is Required to Mediate Nuclear Import of Ran-Gdp
J.Mol.Biol., 293, 1999
|
|
1QLG
 
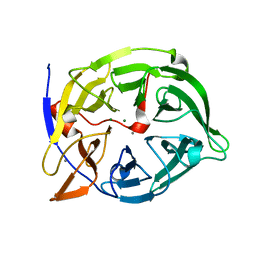 | | Crystal structure of phytase with magnesium from Bacillus amyloliquefaciens | | Descriptor: | 3-PHYTASE, CALCIUM ION, MAGNESIUM ION | | Authors: | Shin, S, Ha, N.-C, Oh, B.-H. | | Deposit date: | 1999-08-31 | | Release date: | 2000-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of a Novel, Thermostable Phytase in Partially and Fully Calcium-Loaded States
Nat.Struct.Biol., 7, 2000
|
|
1QM5
 
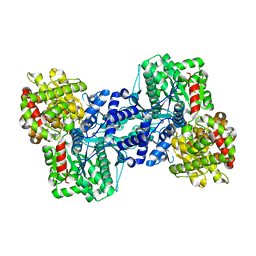 | | Phosphorylase recognition and phosphorylysis of its oligosaccharide substrate: answers to a long outstanding question | | Descriptor: | 4-thio-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, MALTODEXTRIN PHOSPHORYLASE, PHOSPHATE ION, ... | | Authors: | Watson, K.A, McCleverty, C, Geremia, S, Cottaz, S, Driguez, H, Johnson, L.N. | | Deposit date: | 1999-09-20 | | Release date: | 2000-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylase Recognition and Phosphorolysis of its Oligosaccharide Substrate: Answers to a Long Outstanding Question
Embo J., 18, 1999
|
|
3NOS
 
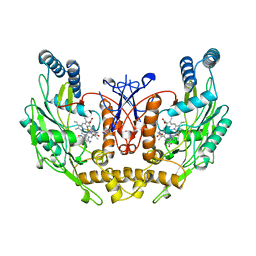 | | HUMAN ENDOTHELIAL NITRIC OXIDE SYNTHASE WITH ARGININE SUBSTRATE | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ENDOTHELIAL NITRIC-OXIDE SYNTHASE, N-OMEGA-HYDROXY-L-ARGININE, ... | | Authors: | Fischmann, T.O, Weber, P.C. | | Deposit date: | 1999-02-03 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of nitric oxide synthase isoforms reveals striking active-site conservation.
Nat.Struct.Biol., 6, 1999
|
|
1DUZ
 
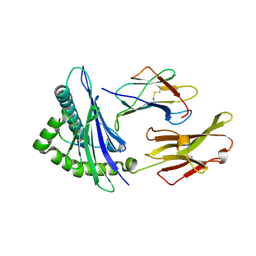 | | HUMAN CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A 0201) IN COMPLEX WITH A NONAMERIC PEPTIDE FROM HTLV-1 TAX PROTEIN | | Descriptor: | BETA-2 MICROGLOBULIN, HLA-A*0201, HTLV-1 OCTAMERIC TAX PEPTIDE | | Authors: | Khan, A.R, Baker, B.M, Ghosh, P, Biddison, W.E, Wiley, D.C. | | Deposit date: | 2000-01-19 | | Release date: | 2000-02-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure and stability of an HLA-A*0201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site.
J.Immunol., 164, 2000
|
|
1DYM
 
 | | Humicola insolens Endocellulase Cel7B (EG 1) E197A Mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Davies, G.J, Moraz, O, Driguez, H, Schulein, M. | | Deposit date: | 2000-02-03 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the family 7 endoglucanase I (Cel7B) from Humicola insolens at 2.2 A resolution and identification of the catalytic nucleophile by trapping of the covalent glycosyl-enzyme intermediate.
Biochem.J., 335 ( Pt 2), 1998
|
|
1DUY
 
 | | CRYSTAL STRUCTURE OF HLA-A*0201/OCTAMERIC TAX PEPTIDE COMPLEX | | Descriptor: | BETA-2 MICROGLOBULIN, HLA-A2*0201, HTLV-1 OCTAMERIC TAX PEPTIDE | | Authors: | Khan, A.R, Baker, B.M, Ghosh, P, Biddison, W.E, Wiley, D.C. | | Deposit date: | 2000-01-19 | | Release date: | 2000-02-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure and stability of an HLA-A*0201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site.
J.Immunol., 164, 2000
|
|
1QNK
 
 | | TRUNCATED HUMAN GROB[5-73], NMR, 20 STRUCTURES | | Descriptor: | C-X-C motif chemokine 2 | | Authors: | Qian, Y.Q, Johanson, K, McDevitt, P. | | Deposit date: | 1999-10-18 | | Release date: | 2000-02-04 | | Last modified: | 2018-06-13 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of truncated human GRObeta [5-73] and its structural comparison with CXC chemokine family members GROalpha and IL-8.
J. Mol. Biol., 294, 1999
|
|
1CQ1
 
 | | Soluble Quinoprotein Glucose Dehydrogenase from Acinetobacter Calcoaceticus in Complex with PQQH2 and Glucose | | Descriptor: | CALCIUM ION, PYRROLOQUINOLINE QUINONE, SOLUBLE QUINOPROTEIN GLUCOSE DEHYDROGENASE, ... | | Authors: | Oubrie, A, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1999-08-04 | | Release date: | 2000-02-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of soluble quinoprotein glucose dehydrogenase.
EMBO J., 18, 1999
|
|
4NOS
 
 | | HUMAN INDUCIBLE NITRIC OXIDE SYNTHASE WITH INHIBITOR | | Descriptor: | 2-AMINO-6-(1,2-DIHYDROXY-PROPYL)-7,8-DIHYDRO-6H-PTERIDIN-4-ONE, 5,6,7,8-TETRAHYDROBIOPTERIN, ETHYLISOTHIOUREA, ... | | Authors: | Fischmann, T.O, Weber, P.C. | | Deposit date: | 1999-02-03 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural characterization of nitric oxide synthase isoforms reveals striking active-site conservation.
Nat.Struct.Biol., 6, 1999
|
|
1NPS
 
 | | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF PROTEIN S | | Descriptor: | CALCIUM ION, DEVELOPMENT-SPECIFIC PROTEIN S | | Authors: | Wenk, M, Baumgartner, R, Mayer, E.M, Huber, R, Holak, T.A, Jaenicke, R. | | Deposit date: | 1999-02-01 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The domains of protein S from Myxococcus xanthus: structure, stability and interactions.
J.Mol.Biol., 286, 1999
|
|
1QMT
 
 | |
1ED0
 
 | | NMR structural determination of viscotoxin A3 from Viscum album L. | | Descriptor: | VISCOTOXIN A3 | | Authors: | Romagnoli, S, Ugolini, R, Fogolari, F, Schaller, G, Urech, K, Giannattasio, M, Ragona, L, Molinari, H. | | Deposit date: | 2000-01-26 | | Release date: | 2000-02-04 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structural determination of viscotoxin A3 from Viscum album L.
Biochem.J., 350, 2000
|
|
2HLP
 
 | | CRYSTAL STRUCTURE OF THE E267R MUTANT OF A HALOPHILIC MALATE DEHYDROGENASE IN THE APO FORM | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE, SODIUM ION | | Authors: | Richard, S.B, Madern, D, Garcin, E, Zaccai, G. | | Deposit date: | 1999-04-23 | | Release date: | 2000-02-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Halophilic adaptation: novel solvent protein interactions observed in the 2.9 and 2.6 A resolution structures of the wild type and a mutant of malate dehydrogenase from Haloarcula marismortui.
Biochemistry, 39, 2000
|
|
