2M7P
 
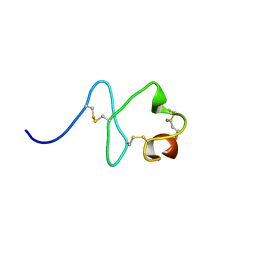 | | RXFP1 utilises hydrophobic moieties on a signalling surface of the LDLa module to mediate receptor activation | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor, Relaxin receptor 1 | | Authors: | Kong, R.CK, Petrie, E.J, Mohanty, B, Ling, J, Lee, J.C.Y, Gooley, P.R, Bathgate, R.A.D. | | Deposit date: | 2013-04-29 | | Release date: | 2013-08-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The relaxin receptor (RXFP1) utilizes hydrophobic moieties on a signaling surface of its N-terminal low density lipoprotein class A module to mediate receptor activation.
J.Biol.Chem., 288, 2013
|
|
1FP2
 
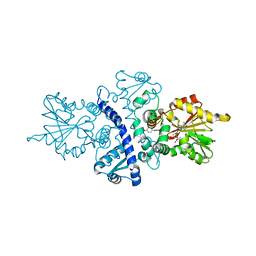 | | CRYSTAL STRUCTURE ANALYSIS OF ISOFLAVONE O-METHYLTRANSFERASE | | Descriptor: | 4'-HYDROXY-7-METHOXYISOFLAVONE, ISOFLAVONE O-METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zubieta, C, Dixon, R.A, Noel, J.P. | | Deposit date: | 2000-08-29 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases.
Nat.Struct.Biol., 8, 2001
|
|
1RKJ
 
 | | Solution structure of the complex formed by the two N-terminal RNA-binding domains of nucleolin and a pre-rRNA target | | Descriptor: | 5'-R(*GP*GP*AP*UP*GP*CP*CP*UP*CP*CP*CP*GP*AP*GP*UP*GP*CP*AP*UP*CP*C)-3', Nucleolin | | Authors: | Johansson, C, Finger, L.D, Trantirek, L, Mueller, T.D, Kim, S, Laird-Offringa, I.A, Feigon, J. | | Deposit date: | 2003-11-21 | | Release date: | 2004-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the complex formed by the two N-terminal RNA-binding domains of nucleolin and a pre-rRNA target.
J.Mol.Biol., 337, 2004
|
|
8CIQ
 
 | | JzTx-34 toxin peptide | | Descriptor: | Mu-theraphotoxin-Cg1a | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2023-02-10 | | Release date: | 2023-07-26 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of new peptides activating human Na v 1.1.
Biomed Pharmacother, 165, 2023
|
|
8CJP
 
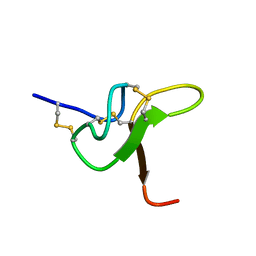 | | JzTx-34 toxin peptide H18A mutant | | Descriptor: | Mu-theraphotoxin-Cg1a | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of new peptides activating human Na v 1.1.
Biomed Pharmacother, 165, 2023
|
|
8CJS
 
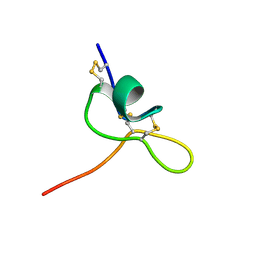 | | JzTx-34 toxin peptide W31A mutant | | Descriptor: | Mu-theraphotoxin-Cg1a | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of new peptides activating human Na v 1.1.
Biomed Pharmacother, 165, 2023
|
|
8CJQ
 
 | | JzTx-34 toxin peptide E20A mutant | | Descriptor: | Mu-theraphotoxin-Cg1a | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of new peptides activating human Na v 1.1.
Biomed Pharmacother, 165, 2023
|
|
8CJR
 
 | | JzTx-34 toxin peptide W25A mutant | | Descriptor: | Mu-theraphotoxin-Cg1a | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of new peptides activating human Na v 1.1.
Biomed Pharmacother, 165, 2023
|
|
8CJT
 
 | | JzTx-34 toxin peptide W33A mutant | | Descriptor: | Mu-theraphotoxin-Cg1a | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of new peptides activating human Na v 1.1.
Biomed Pharmacother, 165, 2023
|
|
3APZ
 
 | |
3UP3
 
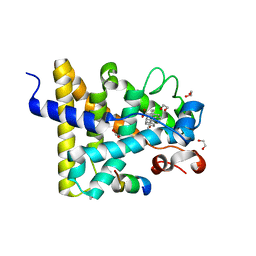 | | Nuclear receptor DAF-12 from hookworm Ancylostoma ceylanicum in complex with (25S)-cholestenoic acid | | Descriptor: | (8alpha,10alpha,25S)-3-hydroxycholesta-3,5-dien-26-oic acid, 1,2-ETHANEDIOL, Nuclear receptor coactivator 2, ... | | Authors: | Zhi, X, Zhou, X.E, Melcher, K, Motola, D.L, Gelmedin, V, Hawdon, J, Kliewer, S.A, Mangelsdorf, D.J, Xu, H.E. | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Conservation of Ligand Binding Reveals a Bile Acid-like Signaling Pathway in Nematodes.
J.Biol.Chem., 287, 2012
|
|
1EDP
 
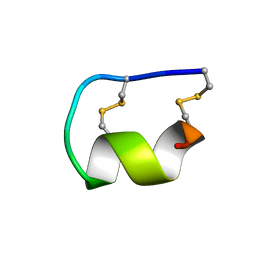 | |
4NY5
 
 | |
1FGA
 
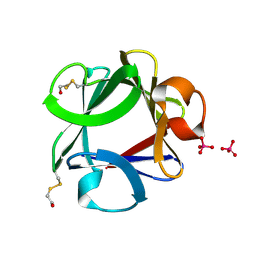 | |
299D
 
 | | CAPTURING THE STRUCTURE OF A CATALYTIC RNA INTERMEDIATE: THE HAMMERHEAD RIBOZYME | | Descriptor: | RNA HAMMERHEAD RIBOZYME | | Authors: | Scott, W.G, Murray, J.B, Arnold, J.R.P, Stoddard, B.L, Klug, A. | | Deposit date: | 1996-12-14 | | Release date: | 1997-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme.
Science, 274, 1996
|
|
2QD5
 
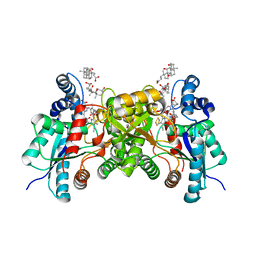 | | Structure of wild type human ferrochelatase in complex with a lead-porphyrin compound | | Descriptor: | ACETIC ACID, CHOLIC ACID, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Meldock, A.E, Dailey, T.A, Ross, T.A, Dailey, H.A, Lanzilotta, W.N. | | Deposit date: | 2007-06-20 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A pi-Helix Switch Selective for Porphyrin Deprotonation and Product Release in Human Ferrochelatase.
J.Mol.Biol., 373, 2007
|
|
5DWX
 
 | |
1TQH
 
 | | Covalent Reaction intermediate Revealed in Crystal Structure of the Geobacillus stearothermophilus Carboxylesterase Est30 | | Descriptor: | Carboxylesterase precursor, PROPYL ACETATE, SULFATE ION | | Authors: | Liu, P, Wang, Y.F, Ewis, H.E, Abdelal, A.T, Lu, C.D, Harrison, R.W, Weber, I.T. | | Deposit date: | 2004-06-17 | | Release date: | 2004-09-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Covalent reaction intermediate revealed in crystal structure of the Geobacillus stearothermophilus carboxylesterase Est30.
J.Mol.Biol., 342, 2004
|
|
1JX1
 
 | | Chalcone Isomerase--T48A mutant | | Descriptor: | 7-HYDROXY-2-(4-HYDROXY-PHENYL)-CHROMAN-4-ONE, CHALCONE--FLAVONONE ISOMERASE 1, SULFATE ION | | Authors: | Jez, J.M, Bowman, M.E, Noel, J.P. | | Deposit date: | 2001-09-05 | | Release date: | 2002-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of Hydrogen Bonds in the Reaction Mechanism of Chalcone Isomerase
Biochemistry, 41, 2002
|
|
3ZUG
 
 | | E268D mutant of FAD synthetase from Corynebacterium ammoniagenes | | Descriptor: | RIBOFLAVIN BIOSYNTHESIS PROTEIN RIBF, SULFATE ION | | Authors: | Herguedas, B, Martinez-Julvez, M, Serrano, A, Medina, M. | | Deposit date: | 2011-07-19 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Key Residues at the Riboflavin Kinase Catalytic Site of the Bifunctional Riboflavin Kinase/Fmn Adenylyltransferase from Corynebacterium Ammoniagenes.
Cell Biochem.Biophys., 65, 2013
|
|
5DWW
 
 | |
1XVL
 
 | | The three-dimensional structure of MntC from Synechocystis 6803 | | Descriptor: | MANGANESE (II) ION, Mn transporter | | Authors: | Rukhman, V, Anati, R, Melamed-Frank, M, Bhattacharyya-Pakrasi, M, Pakrasi, H.B, Adir, N. | | Deposit date: | 2004-10-28 | | Release date: | 2005-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The MntC crystal structure suggests that import of Mn2+ in cyanobacteria is redox controlled.
J.Mol.Biol., 348, 2005
|
|
1JX0
 
 | | Chalcone Isomerase--Y106F mutant | | Descriptor: | 7-HYDROXY-2-(4-HYDROXY-PHENYL)-CHROMAN-4-ONE, CHALCONE--FLAVONONE ISOMERASE 1, SULFATE ION | | Authors: | Jez, J.M, Bowman, M.E, Noel, J.P. | | Deposit date: | 2001-09-05 | | Release date: | 2002-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Role of Hydrogen Bonds in the Reaction Mechanism of Chalcone Isomerase
Biochemistry, 41, 2002
|
|
7D84
 
 | | 34-fold symmetry Salmonella S ring formed by full-length FliF | | Descriptor: | Flagellar M-ring protein | | Authors: | Kawamoto, A, Miyata, T, Makino, F, Kinoshita, M, Minamino, T, Imada, K, Kato, T, Namba, K. | | Deposit date: | 2020-10-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Native flagellar MS ring is formed by 34 subunits with 23-fold and 11-fold subsymmetries.
Nat Commun, 12, 2021
|
|
6T3O
 
 | | Crystal structure of the human myomesin domain 10 | | Descriptor: | AZIDE ION, Myomesin-1 | | Authors: | Duskova, J, Petrokova, H, Maly, P. | | Deposit date: | 2019-10-11 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Myomedin scaffold variants targeted to 10E8 HIV-1 broadly neutralizing antibody mimic gp41 epitope and elicit HIV-1 virus-neutralizing sera in mice.
Virulence, 12, 2021
|
|
