1H37
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, SQUALENE--HOPENE CYCLASE, {4-[((1S,2S)-2-{[ALLYL(CYCLOPROPYL)AMINO]METHYL}CYCLOPROPYL)METHOXY]PHENYL}(4-BROMOPHENYL)METHANONE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-24 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
1H38
 
 | | Structure of a T7 RNA polymerase elongation complex at 2.9A resolution | | Descriptor: | 5'-D(*GP*GP*GP*AP*AP*TP*CP*GP*AP*CP *AP*TP*CP*GP*CP*CP*GP*C)-3', 5'-D(*GP*TP*CP*GP*AP*TP*TP*CP*CP*CP)-3', 5'-R(*AP*AP*CP*UP*GP*CP*GP*GP*CP*GP *AP*U)-3', ... | | Authors: | Tahirov, T.H, Temyakov, D, Anikin, M, Patlan, V, McAllister, W.T, Vassylyev, D.G, Yokoyama, S. | | Deposit date: | 2002-08-24 | | Release date: | 2002-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a T7 RNA Polymerase Elongation Complex at 2.9 A Resolution
Nature, 420, 2002
|
|
1H39
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, ALLYL-{6-[3-(4-BROMO-PHENYL)-1-METHYL-1H-INDAZOL-6-YL]OXY}HEXYL)-N-METHYLAMINE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-24 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
1H3A
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (2E)-N-ALLYL-4-{[3-(4-BROMOPHENYL)-5-FLUORO-1-METHYL-1H-INDAZOL-6-YL]OXY}-N-METHYL-2-BUTEN-1-AMINE, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-24 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
1H3B
 
 | | Squalene-Hopene Cyclase | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, N-{6-[4-(6-BROMO-1,2-BENZISOTHIAZOL-3-YL)PHENOXY]HEXYL}-N-METHYL-2-PROPEN-1-AMINE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-25 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
1H3C
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, N-(6-{[3-(4-BROMOPHENYL)-1,2-BENZISOTHIAZOL-6-YL]OXY}HEXYL)-N-METHYLPROP-2-EN-1-AMINE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-25 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
1H3D
 
 | | STRUCTURE OF THE E.COLI ATP-PHOSPHORIBOSYLTRANSFERASE | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-PHOSPHORIBOSYLTRANSFERASE, L(+)-TARTARIC ACID | | Authors: | Lohkamp, B, McDermott, G, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-08-27 | | Release date: | 2003-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of Escherichia Coli ATP-Phosphoribosyltransferase: Identification of Substrate Binding Sites and Mode of AMP Inhibition
J.Mol.Biol., 336, 2004
|
|
1H3E
 
 | | Tyrosyl-tRNA synthetase from Thermus thermophilus complexed with wild-type tRNAtyr(GUA) and with ATP and tyrosinol | | Descriptor: | 4-[(2S)-2-amino-3-hydroxypropyl]phenol, ADENOSINE-5'-TRIPHOSPHATE, TYROSYL-TRNA SYNTHETASE, ... | | Authors: | Cusack, S, Yaremchuk, A, Kriklivyi, I, Tukalo, M. | | Deposit date: | 2002-08-28 | | Release date: | 2002-10-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Class I Tyrosyl-tRNA Synthetase Has a Class II Mode or tRNA Recognition
Embo J., 21, 2002
|
|
1H3F
 
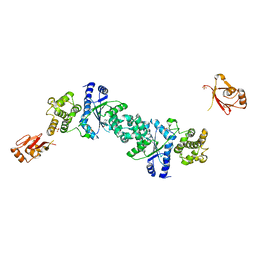 | | Tyrosyl-tRNA synthetase from Thermus thermophilus complexed with tyrosinol | | Descriptor: | 4-[(2S)-2-amino-3-hydroxypropyl]phenol, SULFATE ION, TYROSYL-TRNA SYNTHETASE | | Authors: | Cusack, S, Yaremchuk, A, Kriklivyi, I, Tukalo, M. | | Deposit date: | 2002-08-28 | | Release date: | 2002-09-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Class I Tyrosyl-tRNA Synthetase Has a Class II Mode or tRNA Recognition
Embo J., 21, 2002
|
|
1H3G
 
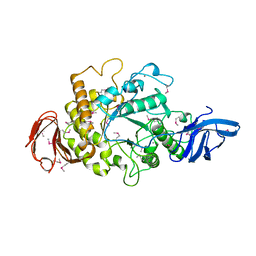 | | Cyclomaltodextrinase from Flavobacterium sp. No. 92: from DNA sequence to protein structure | | Descriptor: | CALCIUM ION, Cyclomaltodextrinase | | Authors: | Fritzsche, H.B, Schwede, T, Jelakovic, S, Schulz, G.E. | | Deposit date: | 2002-09-03 | | Release date: | 2003-08-14 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Covalent and Three-Dimensional Structure of the Cyclodextrinase from Flavobacterium Sp. No. 92.
Eur.J.Biochem., 270, 2003
|
|
1H3H
 
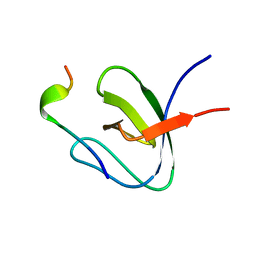 | | Structural Basis for Specific Recognition of an RxxK-containing SLP-76 peptide by the Gads C-terminal SH3 domain | | Descriptor: | GRB2-RELATED ADAPTOR PROTEIN 2, LYMPHOCYTE CYTOSOLIC PROTEIN 2 | | Authors: | Liu, Q, Berry, D, Nash, P, Pawson, T, McGlade, C.J, Li, S.S. | | Deposit date: | 2002-09-03 | | Release date: | 2003-03-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Specific Binding of the Gads SH3 Domain to an Rxxk Motif-Containing Slp-76 Peptide: A Novel Mode of Peptide Recognition
Mol.Cell, 11, 2003
|
|
1H3I
 
 | | Crystal structure of the Histone Methyltransferase SET7/9 | | Descriptor: | HISTONE H3 LYSINE 4 SPECIFIC METHYLTRANSFERASE, MAGNESIUM ION | | Authors: | Wilson, J.R, Jing, C, Walker, P.A, Martin, S.R, Howell, S.A, Blackburn, G.M, Gamblin, S.J, Xiao, B. | | Deposit date: | 2002-09-04 | | Release date: | 2002-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Functional Analysis of the Histone Methyltransferase Set7/9
Cell(Cambridge,Mass.), 111, 2002
|
|
1H3J
 
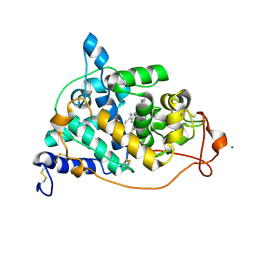 | | STRUCTURE OF RECOMBINANT COPRINUS CINEREUS PEROXIDASE DETERMINED TO 2.0 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Petersen, J.F.W, Houborg, K, Harris, P, Larsen, S. | | Deposit date: | 2002-09-05 | | Release date: | 2003-06-12 | | Last modified: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Impact of the Physical and Chemical Environment on the Molecular Structure of Coprinus Cinereus Peroxidase
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1H3L
 
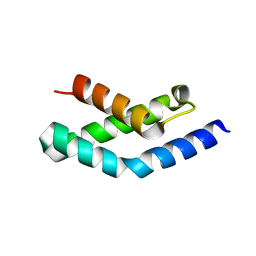 | | N-terminal fragment of SigR from Streptomyces coelicolor | | Descriptor: | RNA POLYMERASE SIGMA FACTOR | | Authors: | Li, W, Stevenson, C.E.M, Burton, N, Jakimowicz, P, Paget, M.S.B, Buttner, M.J, Lawson, D.M, Kleanthous, C. | | Deposit date: | 2002-09-10 | | Release date: | 2002-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.375 Å) | | Cite: | Identification and Structure of the Anti-Sigma Factor-Binding Domain of the Disulfide-Stress Regulated Sigma Factor Sigma(R) from Streptomyces Coelicolor
J.Mol.Biol., 323, 2002
|
|
1H3M
 
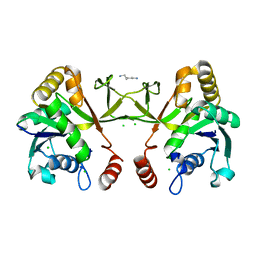 | | Structure of 4-diphosphocytidyl-2C-methyl-D-erythritol synthetase | | Descriptor: | 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLYLTRANSFERASE, CHLORIDE ION, PENTANE-1,5-DIAMINE | | Authors: | Kemp, L.E, Bond, C.S, Hunter, W.N. | | Deposit date: | 2002-09-10 | | Release date: | 2003-08-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Tetragonal Crystal Form of Escherichia Coli 2-C-Methyl-D-Erythritol 4-Phosphate Cytidylyltransferase
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1H3N
 
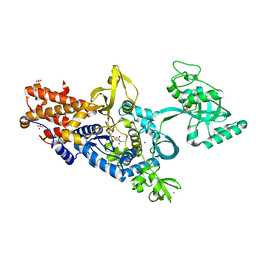 | | Leucyl-tRNA synthetase from Thermus thermophilus complexed with a sulphamoyl analogue of leucyl-adenylate | | Descriptor: | LEUCINE, LEUCYL-TRNA SYNTHETASE, SULFATE ION, ... | | Authors: | Cusack, S, Yaremchuk, A, Tukalo, M. | | Deposit date: | 2002-09-11 | | Release date: | 2002-09-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2A Structure of Leucyl-tRNA Synthetase and its Complex with a Leucyl-Adenylate Analogue
Embo J., 19, 2000
|
|
1H3O
 
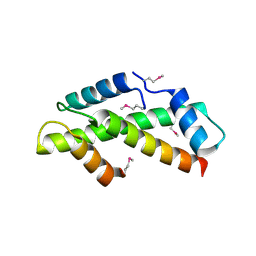 | | Crystal Structure of the Human TAF4-TAF12 (TAFII135-TAFII20) Complex | | Descriptor: | TRANSCRIPTION INITIATION FACTOR TFIID 135 KDA SUBUNIT, TRANSCRIPTION INITIATION FACTOR TFIID 20/15 KDA SUBUNITS | | Authors: | Werten, S, Mitschler, A, Moras, D. | | Deposit date: | 2002-09-12 | | Release date: | 2002-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a Subcomplex of Human Transcription Factor TFIID Formed by TATA Binding Protein-Associated Factors Htaf4 (Htaf(II)135) and Htaf12 (Htaf(II)20).
J.Biol.Chem., 277, 2002
|
|
1H3P
 
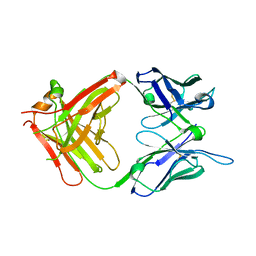 | | STRUCTURAL CHARACTERISATION OF A MONOCLONAL ANTIBODY SPECIFIC FOR THE PRES1 REGION OF THE HEPATITIS B VIRUS | | Descriptor: | ANTIBODY FAB FRAGMENT | | Authors: | Pizarro, J.C, Vulliez-Le-normand, B, Riottot, M.M, Budkowska, A, Bentley, G.A. | | Deposit date: | 2002-09-12 | | Release date: | 2002-09-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Functional Characterisation of a Monoclonal Antibody Specific for the Pres1 Region of Hepatitis B Virus
FEBS Lett., 509, 2001
|
|
1H3Q
 
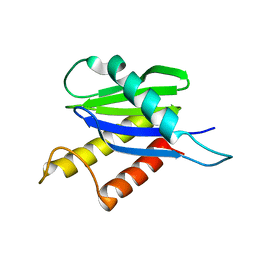 | |
1H3T
 
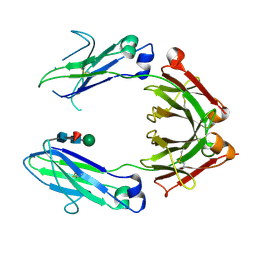 | | Crystal structure of the human igg1 fc-fragment,glycoform (mn2f)2 | | Descriptor: | IG GAMMA-1 CHAIN C REGION, beta-D-mannopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Krapp, S, Mimura, Y, Jefferis, R, Huber, R, Sondermann, P. | | Deposit date: | 2002-09-19 | | Release date: | 2003-01-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analysis of Human Igg-Fc Glycoforms Reveals a Correlation between Glycosylation and Structural Integrity
J.Mol.Biol., 325, 2003
|
|
1H3U
 
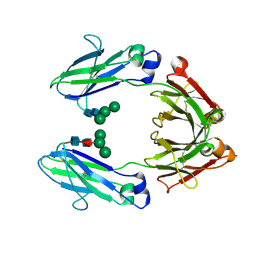 | | CRYSTAL STRUCTURE OF THE HUMAN IGG1 FC-FRAGMENT,GLYCOFORM (M3N2F)2 | | Descriptor: | IG GAMMA-1 CHAIN C REGION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Krapp, S, Mimura, Y, Jefferis, R, Huber, R, Sondermann, P. | | Deposit date: | 2002-09-19 | | Release date: | 2003-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analysis of Human Igg-Fc Glycoforms Reveals a Correlation between Glycosylation and Structural Integrity.
J.Mol.Biol., 325, 2003
|
|
1H3V
 
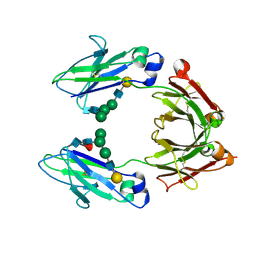 | | CRYSTAL STRUCTURE OF THE HUMAN IGG1 FC-FRAGMENT,GLYCOFORM (G2F)2,SG P212121 | | Descriptor: | IG GAMMA-1 CHAIN C REGION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Krapp, S, Mimura, Y, Jefferis, R, Huber, R, Sondermann, P. | | Deposit date: | 2002-09-19 | | Release date: | 2003-01-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Analysis of Human Igg-Fc Glycoforms Reveals a Correlation between Glycosylation and Structural Integrity
J.Mol.Biol., 325, 2003
|
|
1H3W
 
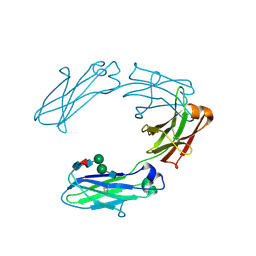 | | Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IG GAMMA-1 CHAIN C REGION | | Authors: | Krapp, S, Mimura, Y, Jefferis, R, Huber, R, Sondermann, P. | | Deposit date: | 2002-09-19 | | Release date: | 2003-01-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Analysis of Human Igg-Fc Glycoforms Reveals a Correlation between Glycosylation and Structural Integrity
J.Mol.Biol., 325, 2003
|
|
1H3X
 
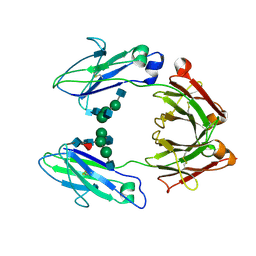 | | CRYSTAL STRUCTURE OF THE HUMAN IGG1 FC-FRAGMENT,GLYCOFORM (G0F)2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IG GAMMA-1 CHAIN C REGION | | Authors: | Krapp, S, Mimura, Y, Jefferis, R, Huber, R, Sondermann, P. | | Deposit date: | 2002-09-19 | | Release date: | 2003-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural Analysis of Human Igg-Fc Glycoforms Reveals a Correlation between Glycosylation and Structural Integrity
J.Mol.Biol., 325, 2003
|
|
1H3Y
 
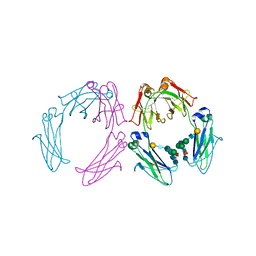 | | Crystal structure of a human IgG1 Fc-fragment,high salt condition | | Descriptor: | IG GAMMA-1 CHAIN C REGION, alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Krapp, S, Mimura, Y, Jefferis, R, Huber, R, Sondermann, P. | | Deposit date: | 2002-09-19 | | Release date: | 2003-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural Analysis of Human Igg-Fc Glycoforms Reveals a Correlation between Glycosylation and Structural Integrity.
J.Mol.Biol., 325, 2003
|
|
