2B3K
 
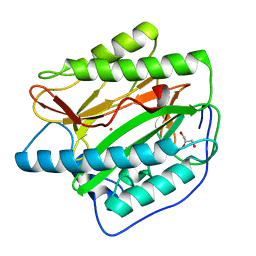 | | Crystal structure of Human Methionine Aminopeptidase Type I in the holo form | | Descriptor: | ACETATE ION, COBALT (II) ION, GLYCEROL, ... | | Authors: | Addlagatta, A, Hu, X, Liu, J.O, Matthews, B.W. | | Deposit date: | 2005-09-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for the Functional Differences between Type I and Type II Human Methionine Aminopeptidases(,).
Biochemistry, 44, 2005
|
|
2LUM
 
 | |
2LVC
 
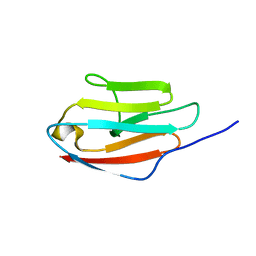 | | Solution NMR Structure of Ig like domain (805-892) of Obscurin-like protein 1 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8578K | | Descriptor: | Obscurin-like protein 1 | | Authors: | Pulavarti, S, Eletsky, A, Sukumaran, D.K, Lee, D, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Pederson, K, Prestegard, J, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-30 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Ig like domain (805-892) of Obscurin-like protein 1 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8578K (CASP Target)
To be Published
|
|
5KYI
 
 | |
5KXV
 
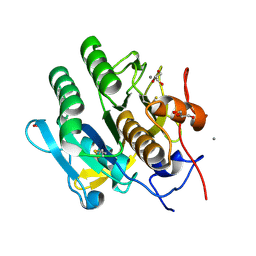 | | Structure Proteinase K at 0.98 Angstroms | | Descriptor: | CALCIUM ION, GLYCEROL, NITRATE ION, ... | | Authors: | Masuda, T, Suzuki, M, Inoue, S, Numata, K, Sugahara, M. | | Deposit date: | 2016-07-20 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic resolution structure of serine protease proteinase K at ambient temperature.
Sci Rep, 7, 2017
|
|
4UD0
 
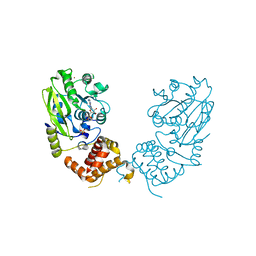 | | X-ray structure and activities of an essential Mononegavirales L- protein domain | | Descriptor: | RNA-DIRECTED RNA POLYMERASE L, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Paesen, G.C, Collet, A, Sallamand, C, Debart, F, Vasseur, J.J, Canard, B, Decroly, E, Grimes, J.M. | | Deposit date: | 2014-12-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-Ray Structure and Activities of an Essential Mononegavirales L-Protein Domain.
Nat.Commun., 6, 2015
|
|
4V4X
 
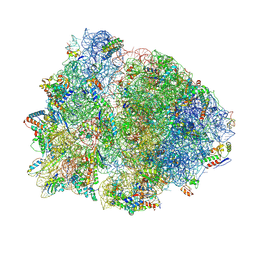 | | Crystal structure of the 70S Thermus thermophilus ribosome showing how the 16S 3'-end mimicks mRNA E and P codons. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Jenner, L, Yusupova, G, Rees, B, Moras, D, Yusupov, M. | | Deposit date: | 2006-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Structural basis for messenger RNA movement on the ribosome.
Nature, 444, 2006
|
|
5L25
 
 | |
5L4L
 
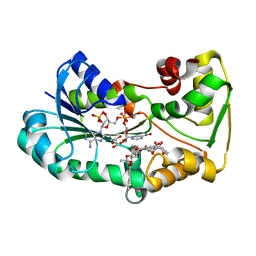 | | polyketide ketoreductase SimC7 - ternary complex with NADP+ and 7-oxo-SD8 | | Descriptor: | 7-oxo-simocyclinone D8, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schafer, M, Stevenson, C.E.M, Wilkinson, B, Lawson, D.M, Buttner, M.J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate-Assisted Catalysis in Polyketide Reduction Proceeds via a Phenolate Intermediate.
Cell Chem Biol, 23, 2016
|
|
8SF8
 
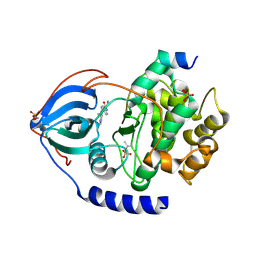 | | Structure of bovine PKA bound to (R)-N-(4-(1H-pyrrolo[2,3-b]pyridin-4-yl)phenyl)-2-amino-4-methylpentanamide | | Descriptor: | N-[4-(1H-pyrrolo[2,3-b]pyridin-4-yl)phenyl]-D-leucinamide, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Coker, J.A, Arya, T, Goins, C.M, Maw, J.J, Macdonald, J.D, Stauffer, S.R. | | Deposit date: | 2023-04-10 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and Characterization of Selective, First-in-Class Inhibitors of Citron Kinase.
J.Med.Chem., 67, 2024
|
|
2AI7
 
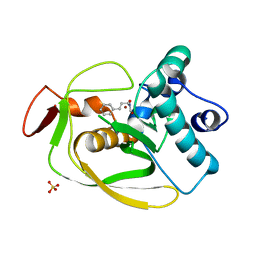 | | S.pneumoniae Polypeptide Deformylase complexed with SB-485345 | | Descriptor: | NICKEL (II) ION, Peptide deformylase, SULFATE ION, ... | | Authors: | Smith, K.J, Petit, C.M, Aubart, K, Smyth, M, McManus, E, Jones, J, Fosberry, A, Lewis, C, Lonetto, M, Christensen, S.B. | | Deposit date: | 2005-07-29 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Variation and inhibitor binding in polypeptide deformylase from four different bacterial species
Protein Sci., 12, 2003
|
|
4UCL
 
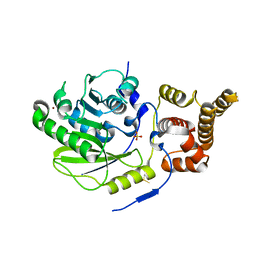 | | X-ray structure and activities of an essential Mononegavirales L- protein domain | | Descriptor: | RNA-DIRECTED RNA POLYMERASE L, SULFATE ION, ZINC ION | | Authors: | Paesen, G.C, Collet, A, Sallamand, C, Debart, F, Vasseur, J.J, Canard, B, Decroly, E, Grimes, J.M. | | Deposit date: | 2014-12-03 | | Release date: | 2015-11-18 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray Structure and Activities of an Essential Mononegavirales L-Protein Domain.
Nat.Commun., 6, 2015
|
|
2AKJ
 
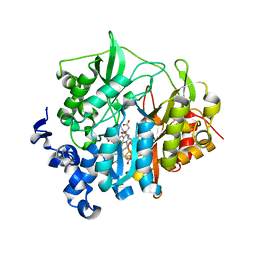 | | Structure of spinach nitrite reductase | | Descriptor: | Ferredoxin--nitrite reductase, chloroplast, IRON/SULFUR CLUSTER, ... | | Authors: | Swamy, U, Wang, M, Tripathy, J.N, Kim, S.-K, Hirasawa, M, Knaff, D.B, Allen, J.P. | | Deposit date: | 2005-08-03 | | Release date: | 2006-01-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Spinach Nitrite Reductase: Implications for Multi-electron Reactions by the Iron-Sulfur:Siroheme Cofactor
Biochemistry, 44, 2005
|
|
2M8C
 
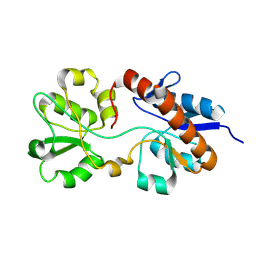 | | The solution NMR structure of E. coli apo-HisJ | | Descriptor: | Cationic amino acid ABC transporter, periplasmic binding protein | | Authors: | Chu, B.C.H, Vogel, H.J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-10-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Role of the Two Structural Domains from the Periplasmic Escherichia coli Histidine-binding Protein HisJ.
J.Biol.Chem., 288, 2013
|
|
4UPH
 
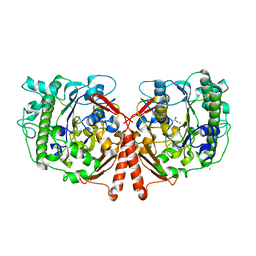 | | Crystal Structure of Phosphonate Monoester Hydrolase of Agrobacterium radiobacter | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SULFATASE (SULFURIC ESTER HYDROLASE) PROTEIN | | Authors: | Fischer, G, Loo, B.v, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2014-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Balancing Specificity and Promiscuity in Enzyme Evolution: Multidimensional Activity Transitions in the Alkaline Phosphatase Superfamily.
J.Am.Chem.Soc., 141, 2019
|
|
2M6L
 
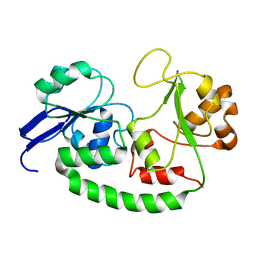 | | Solution structure of the Escherichia coli holo ferric enterobactin binding protein | | Descriptor: | Ferrienterobactin-binding periplasmic protein | | Authors: | Chu, B.C.H, Otten, R, Krewulak, K.D, Mulder, F.A.A, Vogel, H.J. | | Deposit date: | 2013-04-05 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure, binding properties, and dynamics of the bacterial siderophore-binding protein FepB.
J.Biol.Chem., 289, 2014
|
|
5L3Z
 
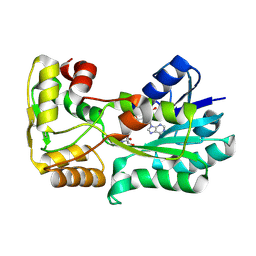 | | polyketide ketoreductase SimC7 - binary complex with NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, polyketide ketoreductase SimC7 | | Authors: | Schafer, M, Stevenson, C.E.M, Wilkinson, B, Lawson, D.M, Buttner, M.J. | | Deposit date: | 2016-05-24 | | Release date: | 2016-10-05 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Substrate-Assisted Catalysis in Polyketide Reduction Proceeds via a Phenolate Intermediate.
Cell Chem Biol, 23, 2016
|
|
4WAL
 
 | | Crystal structure of selenomethionine Msl5 protein in complex with RNA at 2.2 A | | Descriptor: | Branchpoint-bridging protein, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jacewicz, A, Smith, P, Chico, L, Schwer, B, Shuman, S. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for recognition of intron branchpoint RNA by yeast Msl5 and selective effects of interfacial mutations on splicing of yeast pre-mRNAs.
Rna, 21, 2015
|
|
4UID
 
 | |
2AZZ
 
 | | Crystal Structure of Porcine Pancreatic Phospholipase A2 in Complex with Taurocholate | | Descriptor: | CALCIUM ION, CHLORIDE ION, Phospholipase A2, ... | | Authors: | Pan, Y.H, Bahnson, B.J, Jain, M.K. | | Deposit date: | 2005-09-12 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for bile salt inhibition of pancreatic phospholipase A2.
J.Mol.Biol., 369, 2007
|
|
2AUV
 
 | | Solution Structure of HndAc : A Thioredoxin-like [2Fe-2S] Ferredoxin Involved in the NADP-reducing Hydrogenase Complex | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, potential NAD-reducing hydrogenase subunit | | Authors: | Nouailler, M, Morelli, X, Bornet, O, Chetrit, B, Dermoun, Z, Guerlesquin, F. | | Deposit date: | 2005-08-29 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of HndAc: a thioredoxin-like domain involved in the NADP-reducing hydrogenase complex
Protein Sci., 15, 2006
|
|
2B03
 
 | | Crystal Structure of Porcine Pancreatic Phospholipase A2 in Complex with Taurochenodeoxycholate | | Descriptor: | CALCIUM ION, Phospholipase A2, major isoenzyme, ... | | Authors: | Pan, Y.H, Bahnson, B.J, Jain, M.K. | | Deposit date: | 2005-09-12 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for bile salt inhibition of pancreatic phospholipase A2.
J.Mol.Biol., 369, 2007
|
|
2MCV
 
 | | Solid-state NMR structure of piscidin 1 in aligned 1:1 phosphatidylethanolamine/phosphoglycerol lipid bilayers | | Descriptor: | Moronecidin | | Authors: | Fu, R, Tian, Y, Perrin Jr, B.S, Grant, C.V, Hayden, R.M, Pastor, R.W, Cotten, M.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-01-22 | | Last modified: | 2014-03-19 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion.
J.Am.Chem.Soc., 136, 2014
|
|
2B3L
 
 | | Crystal structure of type I human methionine aminopeptidase in the apo form | | Descriptor: | ACETIC ACID, GLYCEROL, Methionine aminopeptidase 1, ... | | Authors: | Addlagatta, A, Hu, X, Liu, J.O, Matthews, B.W. | | Deposit date: | 2005-09-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for the Functional Differences between Type I and Type II Human Methionine Aminopeptidases(,).
Biochemistry, 44, 2005
|
|
2B01
 
 | | Crystal Structure of Porcine Pancreatic Phospholipase A2 in Complex with Taurochenodeoxycholate | | Descriptor: | CALCIUM ION, CHLORIDE ION, Phospholipase A2, ... | | Authors: | Pan, Y.H, Bahnson, B.J, Jain, M.K. | | Deposit date: | 2005-09-12 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for bile salt inhibition of pancreatic phospholipase A2.
J.Mol.Biol., 369, 2007
|
|
