5NMT
 
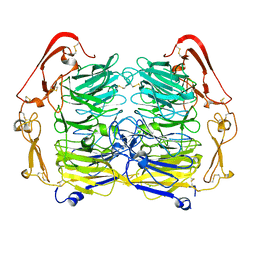 | | Dimer structure of Sortilin ectodomain crystal form 1, 2.3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Leloup, N.O.L, Janssen, B.J.C. | | Deposit date: | 2017-04-07 | | Release date: | 2017-11-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun, 8, 2017
|
|
3FDC
 
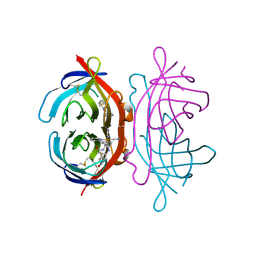 | | Crystal Structure of Avidin | | Descriptor: | Avidin, iron(II) tetracyano-5-(2-Oxo-hexahydro-thieno[3,4-d]imidazol-6-yl)-pentanoic acid (4'-methyl-[2,2']bipyridinyl-4-ylmethyl)-amide | | Authors: | Barker, K.D, Sazinsky, M.H, Eckermann, A.L, Abajian, C, Hartings, M.R, Rosenzweig, A.C, Meade, T.J. | | Deposit date: | 2008-11-25 | | Release date: | 2009-12-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Protein Binding and the Electronic Properties of Iron(II) Complexes: An Electrochemical and Optical Investigation of Outer Sphere Effects.
Bioconjug.Chem., 20, 2009
|
|
5TDD
 
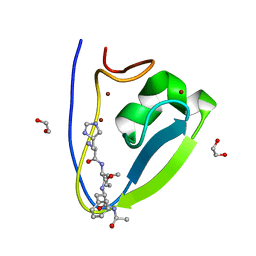 | | Human UBR-box from UBR2 in complex with HIFS peptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UBR2, HIS-ILE-PHE-SER peptide, ... | | Authors: | Munoz-Escobar, J, Kozlov, G, Gehring, K. | | Deposit date: | 2016-09-19 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Bound Waters Mediate Binding of Diverse Substrates to a Ubiquitin Ligase.
Structure, 25, 2017
|
|
9EU6
 
 | | The FK1 domain of FKBP51 in complex with SAFit-analog 23j | | Descriptor: | (1,5-dimethylpyrazol-4-yl)methyl (2~{S})-1-[(2~{S})-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Hausch, F. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-12 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | 1,4-Pyrazolyl-Containing SAFit-Analogues are Selective FKBP51 Inhibitors With Improved Ligand Efficiency and Drug-Like Profile.
Chemmedchem, 19, 2024
|
|
9EUB
 
 | | The FK1 domain of FKBP51 in complex with SAFit-analog 24e | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, [1-(2-hydroxyethyl)pyrazol-4-yl]methyl (2S)-1-[(2S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidine-2-carboxylate | | Authors: | Meyners, C, Buffa, V, Hausch, F. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-12 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1,4-Pyrazolyl-Containing SAFit-Analogues are Selective FKBP51 Inhibitors With Improved Ligand Efficiency and Drug-Like Profile.
Chemmedchem, 19, 2024
|
|
9EU8
 
 | | The FK1 domain of FKBP51 in complex with SAFit-analog 15h | | Descriptor: | (4-methyl-1,3-thiazol-5-yl)methyl (2S)-1-[(2S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Hausch, F. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-12 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 1,4-Pyrazolyl-Containing SAFit-Analogues are Selective FKBP51 Inhibitors With Improved Ligand Efficiency and Drug-Like Profile.
Chemmedchem, 19, 2024
|
|
9EU7
 
 | | The FK1 domain of FKBP51 in complex with SAFit-analog 15b | | Descriptor: | (2-methyl-1,3-thiazol-5-yl)methyl (2S)-1-[(2S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Hausch, F. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-12 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 1,4-Pyrazolyl-Containing SAFit-Analogues are Selective FKBP51 Inhibitors With Improved Ligand Efficiency and Drug-Like Profile.
Chemmedchem, 19, 2024
|
|
9EUC
 
 | | The FK1 domain of FKBP51 in complex with SAFit-analog 23b | | Descriptor: | (1-ethylpyrazol-4-yl)methyl (2S)-1-[(2S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Hausch, F. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-12 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 1,4-Pyrazolyl-Containing SAFit-Analogues are Selective FKBP51 Inhibitors With Improved Ligand Efficiency and Drug-Like Profile.
Chemmedchem, 19, 2024
|
|
5ZNN
 
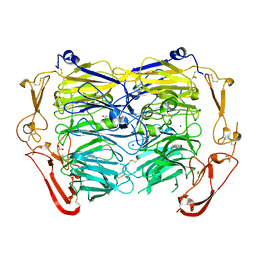 | | Crystal structure of ligand-free form of the Vps10 ectodomain of Sortilin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Yabe-Wada, T, Unno, M. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.448 Å) | | Cite: | Crystal structure of the ligand-free form of the Vps10 ectodomain of dimerized Sortilin at acidic pH
FEBS Lett., 592, 2018
|
|
1V6O
 
 | | Peanut lectin complexed with 10mer peptide (PVRIWSSATG) | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION | | Authors: | Kundhavai Natchiar, S, Arockia Jeyaprakash, A, Ramya, T.N.C, Thomas, C.J, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-12-02 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural plasticity of peanut lectin: an X-ray analysis involving variation in pH, ligand binding and crystal structure.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6ECP
 
 | | The human methylenetetrahydrofolate dehydrogenase/cyclohydrolase (FolD) complexed with NADP and inhibitor LY249543 | | Descriptor: | Methylenetetrahydrofolate dehydrogenase cyclohydrolase, N-(4-{2-[(6S)-2-amino-4-oxo-1,4,5,6,7,8-hexahydropyrido[2,3-d]pyrimidin-6-yl]ethyl}benzoyl)-L-glutamic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bueno, R.V, Dawson, A, Hunter, W.N. | | Deposit date: | 2018-08-08 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An assessment of three human methylenetetrahydrofolate dehydrogenase/cyclohydrolase-ligand complexes following further refinement.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
5OH4
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Piperidine-2,6-dione (Glutarimide) | | Descriptor: | Cereblon isoform 4, ZINC ION, piperidine-2,6-dione | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
5OH2
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Pyrrolidin-2-one (Butyrolactam) | | Descriptor: | Cereblon isoform 4, ZINC ION, pyrrolidin-2-one | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
2B5L
 
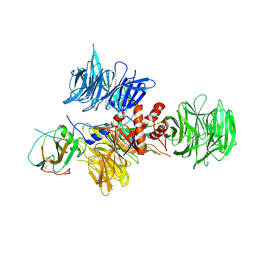 | | Crystal Structure of DDB1 In Complex with Simian Virus 5 V Protein | | Descriptor: | Nonstructural protein V, ZINC ION, damage-specific DNA binding protein 1 | | Authors: | Li, T, Chen, X, Garbutt, K.C, Zhou, P, Zheng, N. | | Deposit date: | 2005-09-28 | | Release date: | 2006-02-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase.
Cell(Cambridge,Mass.), 124, 2006
|
|
4GPO
 
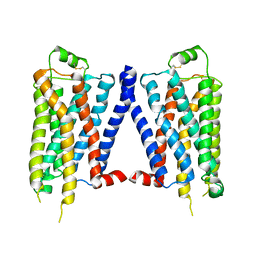 | |
5TDC
 
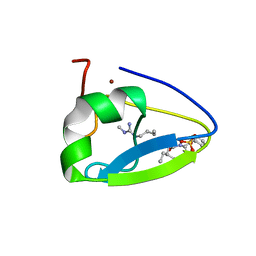 | | Crystal structure of the human UBR-box domain from UBR1 in complex with monomethylated arginine peptide. | | Descriptor: | E3 ubiquitin-protein ligase UBR1, NMM-ILE-PHE-SER peptide, SULFATE ION, ... | | Authors: | Kozlov, G, Munoz-Escobar, J, Matta-Camacho, E, Gehring, K. | | Deposit date: | 2016-09-19 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.607 Å) | | Cite: | Bound Waters Mediate Binding of Diverse Substrates to a Ubiquitin Ligase.
Structure, 25, 2017
|
|
6I2M
 
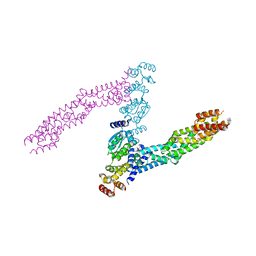 | |
1MKO
 
 | |
4TZU
 
 | | Crystal Structure of Murine Cereblon in Complex with Pomalidomide | | Descriptor: | Protein cereblon, S-Pomalidomide, SULFATE ION, ... | | Authors: | Chamberlain, P.P, Pagarigan, B, Delker, S, Leon, B. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Responsiveness to Thalidomide-Analog Drugs Defined by the Crystal Structure of the Human Cereblon:DDB1:Lenalidomide Complex
to be published
|
|
4J9A
 
 | | Engineered Digoxigenin binder DIG10.3 | | Descriptor: | DIGOXIGENIN, Engineered Digoxigenin binder protein DIG10.3 | | Authors: | Stoddard, B.L, Doyle, L.A. | | Deposit date: | 2013-02-15 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Computational design of ligand-binding proteins with high affinity and selectivity.
Nature, 501, 2013
|
|
4TZC
 
 | | Crystal Structure of Murine Cereblon in Complex with Thalidomide | | Descriptor: | Protein cereblon, S-Thalidomide, SULFATE ION, ... | | Authors: | Chamberlain, P.P, Pagarigan, B, Delker, S, Leon, B. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Basis for Responsiveness to Thalidomide-Analog Drugs Defined by the Crystal Structure of the Human Cereblon:DDB1:Lenalidomide Complex
to be published
|
|
1IRC
 
 | | CYSTEINE RICH INTESTINAL PROTEIN | | Descriptor: | IMIDAZOLE, MYOGLOBIN (METAQUO), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Barrick, D.E, Feese, M. | | Deposit date: | 1995-12-19 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Replacement of the proximal ligand of sperm whale myoglobin with free imidazole in the mutant His-93-->Gly.
Biochemistry, 33, 1994
|
|
3PD9
 
 | | X-ray structure of the ligand-binding core of GluA2 in complex with (R)-5-HPCA at 2.1 A resolution | | Descriptor: | (5R)-3-hydroxy-4,5,6,7-tetrahydroisoxazolo[5,4-c]pyridine-5-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biostructural and pharmacological studies of bicyclic analogues of the 3-isoxazolol glutamate receptor agonist ibotenic acid.
J. Med. Chem., 53, 2010
|
|
2L9M
 
 | | Structure of cIAP1 CARD | | Descriptor: | Baculoviral IAP repeat-containing protein 2 | | Authors: | Day, C.L, Rautureau, G.J.P, Hinds, M.G. | | Deposit date: | 2011-02-21 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | CARD-mediated autoinhibition of cIAP1's E3 ligase activity suppresses cell proliferation and migration.
Mol.Cell, 42, 2011
|
|
3GRR
 
 | |
