7XJ0
 
 | | Structure of human TRPV3 in complex with Trpvicin | | Descriptor: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XIU
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ZINC ION, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-14 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
7XIT
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ZINC ION, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-14 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
7XIS
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | (2-methoxy-4-methoxycarbonyl-phenyl) 5-nitrofuran-2-carboxylate, MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-14 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
7XIK
 
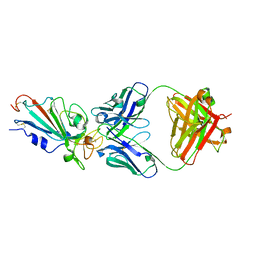 | | SARS-CoV-2-Omicron-RBD and B38-GWP/P-VK antibody complex | | Descriptor: | B38 Fab heavy chain, B38 Fab light chain, Spike protein S1 | | Authors: | Shi, R, Wang, Y, Wu, Z, Han, X, Yan, J. | | Deposit date: | 2022-04-13 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | SARS-CoV-2-Omicron-RBD and B38-GWP/P-VK antibody complex
To Be Published
|
|
7XIJ
 
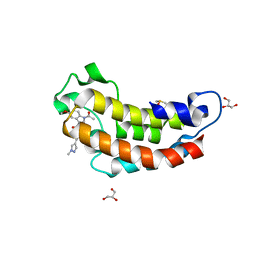 | | Crystal structure of CBP bromodomain liganded with Y08175 | | Descriptor: | 3-[(1-ethanoyl-5-methoxy-indol-3-yl)carbonylamino]-4-fluoranyl-5-(1-methylpyrazol-4-yl)benzoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Xiang, Q, Wang, C, Wu, T, Zhang, Y, Zhang, C, Luo, G, Wu, X, Shen, H, Xu, Y. | | Deposit date: | 2022-04-13 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of CBP bromodomain liganded with Y08175
To Be Published
|
|
7XII
 
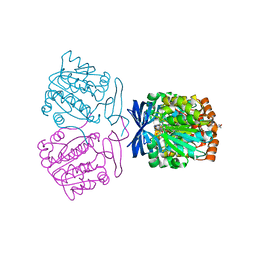 | | Crystal structure of the aminopropyltransferase, SpeE from hyperthermophilic crenarchaeon, Pyrobaculum calidifontis in complex with 5'-methylthioadenosine (MTA) & aminopropylagmatine | | Descriptor: | 1-{4-[(3-aminopropyl)amino]butyl}guanidine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ... | | Authors: | Mizohata, E, Yasuda, Y. | | Deposit date: | 2022-04-13 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate Specificity of an Aminopropyltransferase and the Biosynthesis Pathway of Polyamines in the Hyperthermophilic Crenarchaeon Pyrobaculum calidifontis.
Catalysts, 12, 2022
|
|
7XIH
 
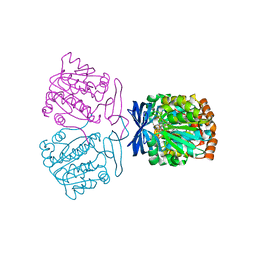 | |
7XIG
 
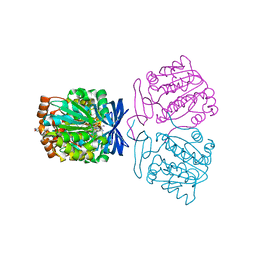 | | Crystal structure of the aminopropyltransferase, SpeE from hyperthermophilic crenarchaeon, Pyrobaculum calidifontis in complex with 5'-methylthioadenosine (MTA) and spermine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Polyamine aminopropyltransferase, ... | | Authors: | Mizohata, E, Yasuda, Y. | | Deposit date: | 2022-04-13 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate Specificity of an Aminopropyltransferase and the Biosynthesis Pathway of Polyamines in the Hyperthermophilic Crenarchaeon Pyrobaculum calidifontis.
Catalysts, 12, 2022
|
|
7XIF
 
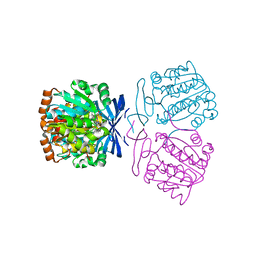 | | Crystal structure of the aminopropyltransferase, SpeE from hyperthermophilic crenarchaeon, Pyrobaculum calidifontis in complex with 5'-methylthioadenosine (MTA) alone or together with spermidine or thermospermine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, N-(3-AMINO-PROPYL)-N-(5-AMINOPROPYL)-1,4-DIAMINOBUTANE, Polyamine aminopropyltransferase, ... | | Authors: | Mizohata, E, Yasuda, Y. | | Deposit date: | 2022-04-13 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Substrate Specificity of an Aminopropyltransferase and the Biosynthesis Pathway of Polyamines in the Hyperthermophilic Crenarchaeon Pyrobaculum calidifontis.
Catalysts, 12, 2022
|
|
7XID
 
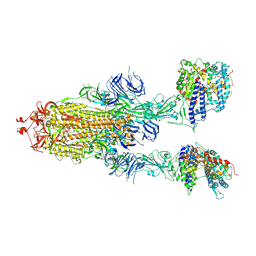 | | S-ECD (Omicron) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2022-04-12 | | Release date: | 2022-06-15 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and functional analysis of an inter-Spike bivalent neutralizing antibody against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7XIC
 
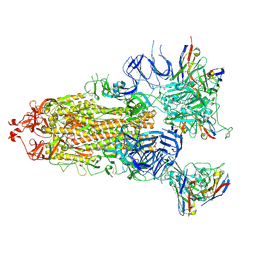 | | S-ECD (Omicron) in complex with STS165 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2022-04-12 | | Release date: | 2022-06-15 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and functional analysis of an inter-Spike bivalent neutralizing antibody against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7XIB
 
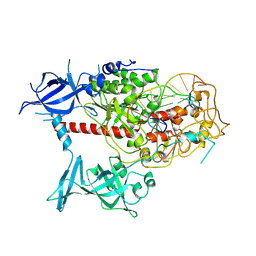 | | Cryo-EM structure of human DNMT1 (aa:351-1616) in complex with ubiquitinated H3 and hemimethylated DNA analog (CXXC-disordered form) | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*AP*(5CM)P*GP*GP*AP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*TP*CP*(C55)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Onoda, H, Kikuchi, A, Kori, S, Yoshimi, S, Yamagata, A, Arita, K. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Structural basis for activation of DNMT1.
Nat Commun, 13, 2022
|
|
7XI9
 
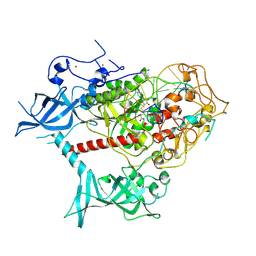 | | Cryo-EM structure of human DNMT1 (aa:351-1616) in complex with ubiquitinated H3 and hemimethylated DNA analog (CXXC-ordered form) | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*AP*(5CM)P*GP*GP*AP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*TP*CP*(C55)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Onoda, H, Kikuchi, A, Kori, S, Yoshimi, S, Yamagata, A, Arita, K. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural basis for activation of DNMT1.
Nat Commun, 13, 2022
|
|
7XI5
 
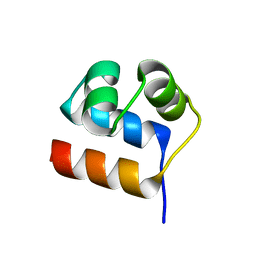 | | Anti-CRISPR-associated Aca10 | | Descriptor: | Transcriptional regulator | | Authors: | Lee, S.Y, Park, H.H. | | Deposit date: | 2022-04-12 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular basis of anti-CRISPR operon repression by Aca10.
Nucleic Acids Res., 50, 2022
|
|
7XI0
 
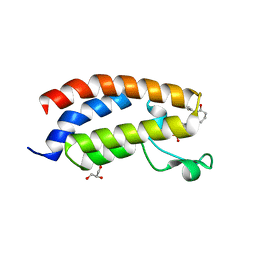 | | Crystal structure of CBP bromodomain liganded with CCS150 | | Descriptor: | (6S)-1-(3-chloranyl-4-methoxy-phenyl)-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(3R)-1-methylsulfonylpyrrolidin-3-yl]benzimidazol-2-yl]piperidin-2-one, CREB-binding protein, GLYCEROL | | Authors: | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-04-11 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
7XHX
 
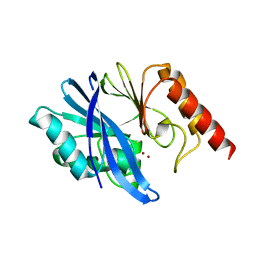 | | Crystal structure of metallo-beta-lactamase IMP-6 | | Descriptor: | Beta-lactamase, ZINC ION | | Authors: | Yamamoto, K, Tanaka, H, Kurisu, G, Nakano, R, Yano, H, Sakai, H. | | Deposit date: | 2022-04-11 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the substrate specificity of IMP-6 and IMP-1 metallo-beta-lactamases.
J.Biochem., 173, 2022
|
|
7XHW
 
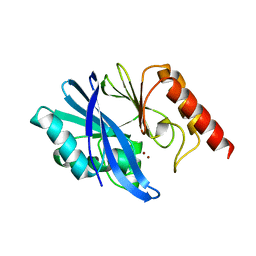 | | Crystal structure of metallo-beta-lactamase IMP-1 | | Descriptor: | Beta-lactamase, ZINC ION | | Authors: | Yamamoto, K, Tanaka, H, Kurisu, G, Nakano, R, Yano, H, Sakai, H. | | Deposit date: | 2022-04-11 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural insights into the substrate specificity of IMP-6 and IMP-1 metallo-beta-lactamases.
J.Biochem., 173, 2022
|
|
7XHU
 
 | | The 0.88 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with tricosanoic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2022-04-10 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | The 0.88 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with tricosanoic acid
To Be Published
|
|
7XHT
 
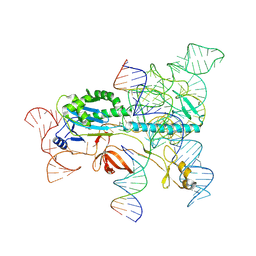 | | Structure of the OgeuIscB-omega RNA-target DNA complex | | Descriptor: | DNA (49-MER), DNA (5'-D(P*GP*AP*AP*GP*AP*AP*AP*AP*CP*CP*AP*T)-3'), LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Kato, K, Okazaki, O, Isayama, Y, Ishikawa, J, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-04-10 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structure of the IscB-omega RNA ribonucleoprotein complex, the likely ancestor of CRISPR-Cas9.
Nat Commun, 13, 2022
|
|
7XHS
 
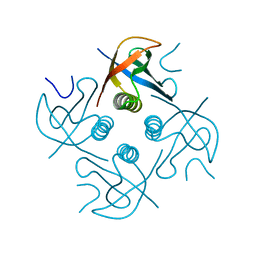 | | Crystal structure of CipA crystal produced by cell-free protein synthesis | | Descriptor: | Cro/Cl family transcriptional regulator | | Authors: | Abe, S, Tanaka, J, Kojima, M, Kanamaru, S, Yamashita, K, Hirata, K, Ueno, T. | | Deposit date: | 2022-04-10 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Cell-free protein crystallization for nanocrystal structure determination.
Sci Rep, 12, 2022
|
|
7XHR
 
 | | Crystal structure of Wild Type Cypovirus Polyhedra produced by cell-free protein synthesis | | Descriptor: | ACETYL GROUP, CHLORIDE ION, Polyhedrin | | Authors: | Abe, S, Tanaka, J, Kojima, M, Hirata, K, Yamashita, K, Ueno, T. | | Deposit date: | 2022-04-10 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Cell-free protein crystallization for nanocrystal structure determination.
Sci Rep, 12, 2022
|
|
7XHM
 
 | | The 0.88 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with behenic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2022-04-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | The 0.88 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with behenic acid
To Be Published
|
|
7XH8
 
 | | The structure of ZCB11 Fab against SARS-CoV-2 Omicron Spike | | Descriptor: | Spike glycoprotein, The heavy chain of ZCB11 antibody, The light chain of ZCB11 antibody | | Authors: | Hang, L, Dang, S. | | Deposit date: | 2022-04-07 | | Release date: | 2022-06-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | A broadly neutralizing antibody protects Syrian hamsters against SARS-CoV-2 Omicron challenge.
Nat Commun, 13, 2022
|
|
7XH6
 
 | | Crystal structure of CBP bromodomain liganded with CCS1477 | | Descriptor: | (6S)-1-[3,4-bis(fluoranyl)phenyl]-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]piperidin-2-one, CREB-binding protein, DIMETHYL SULFOXIDE, ... | | Authors: | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-04-07 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
